Bioinformatics Approach to Identifying Molecular Targets of Isoliquiritigenin Affecting Chronic Obstructive Pulmonary Disease: A Machine Learning Pharmacology Study
Abstract
1. Introduction
2. Results
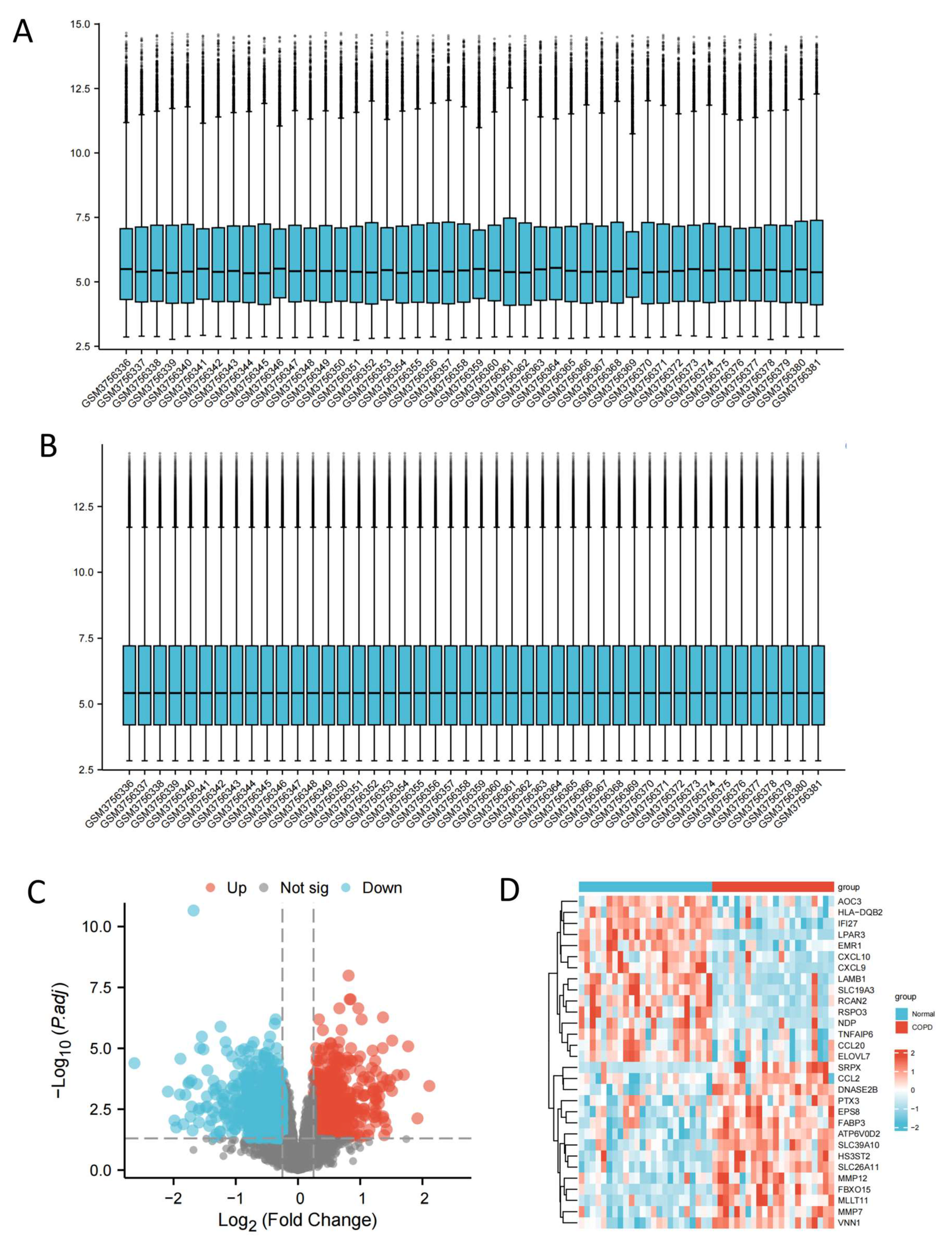
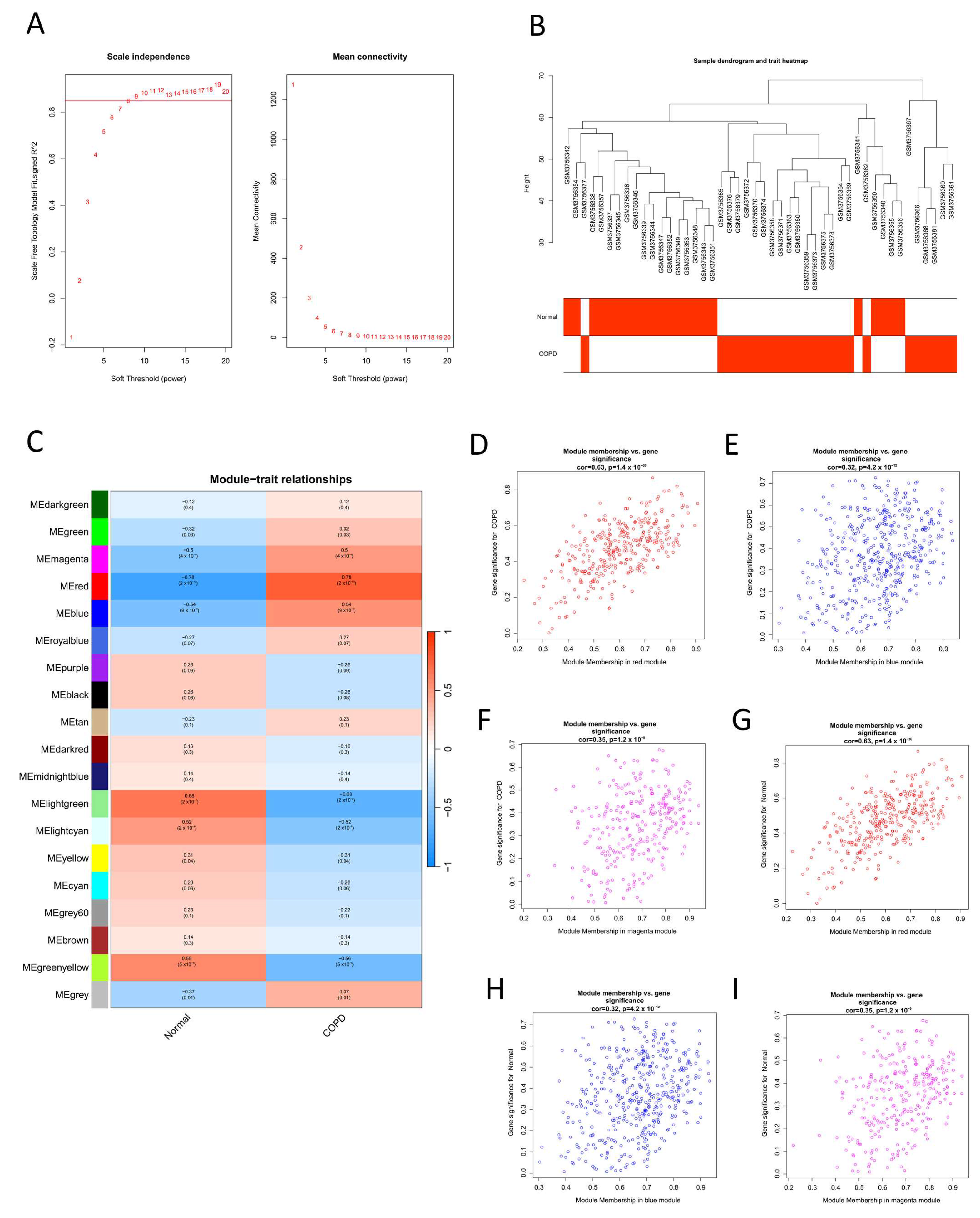
2.1. Identification of COPD-Related Potential Target Genes
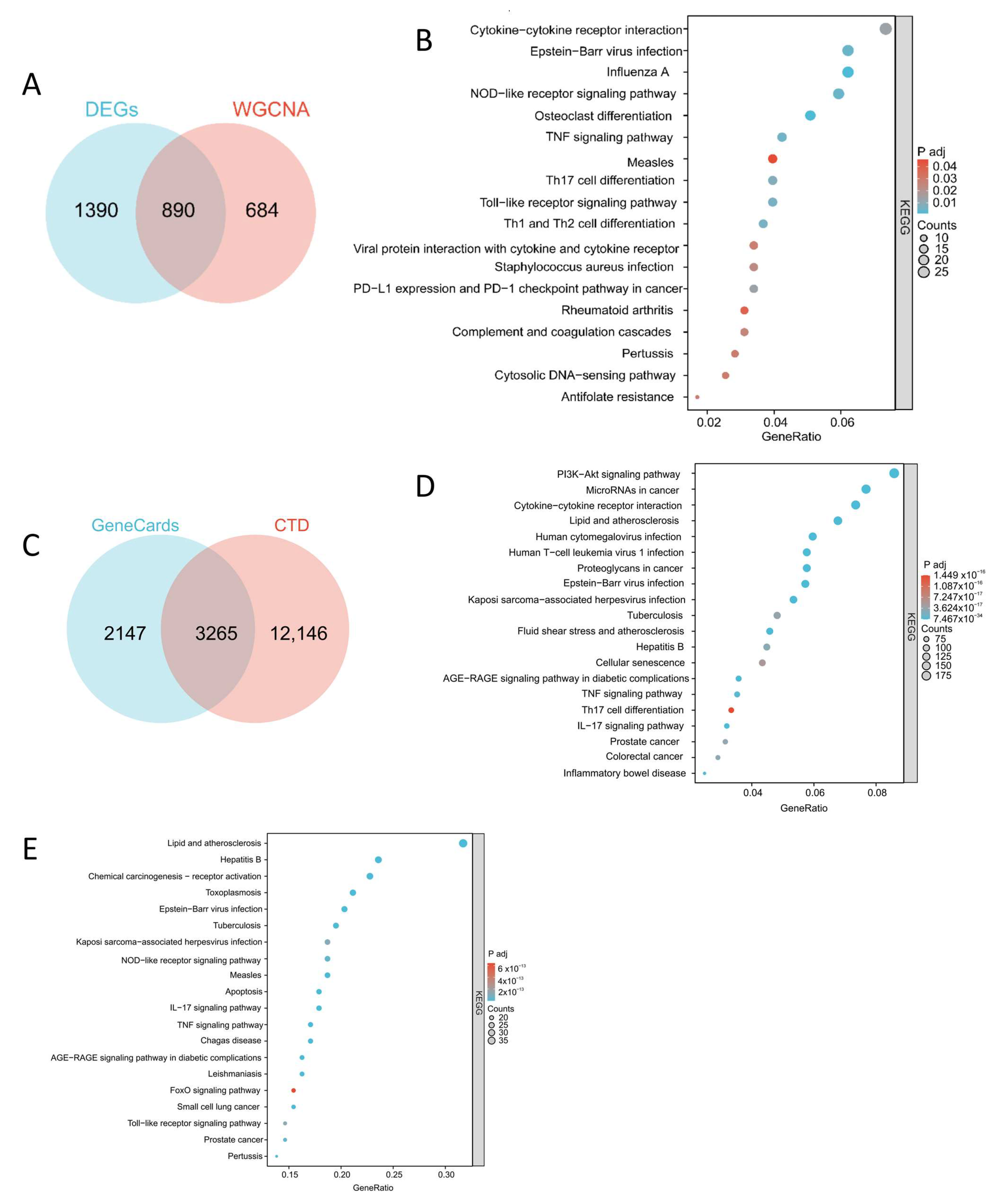
2.2. Construct Drug Small Molecule–Target–Pathway Network and Protein–Protein Interaction (PPI) Network and Screen Key Networks
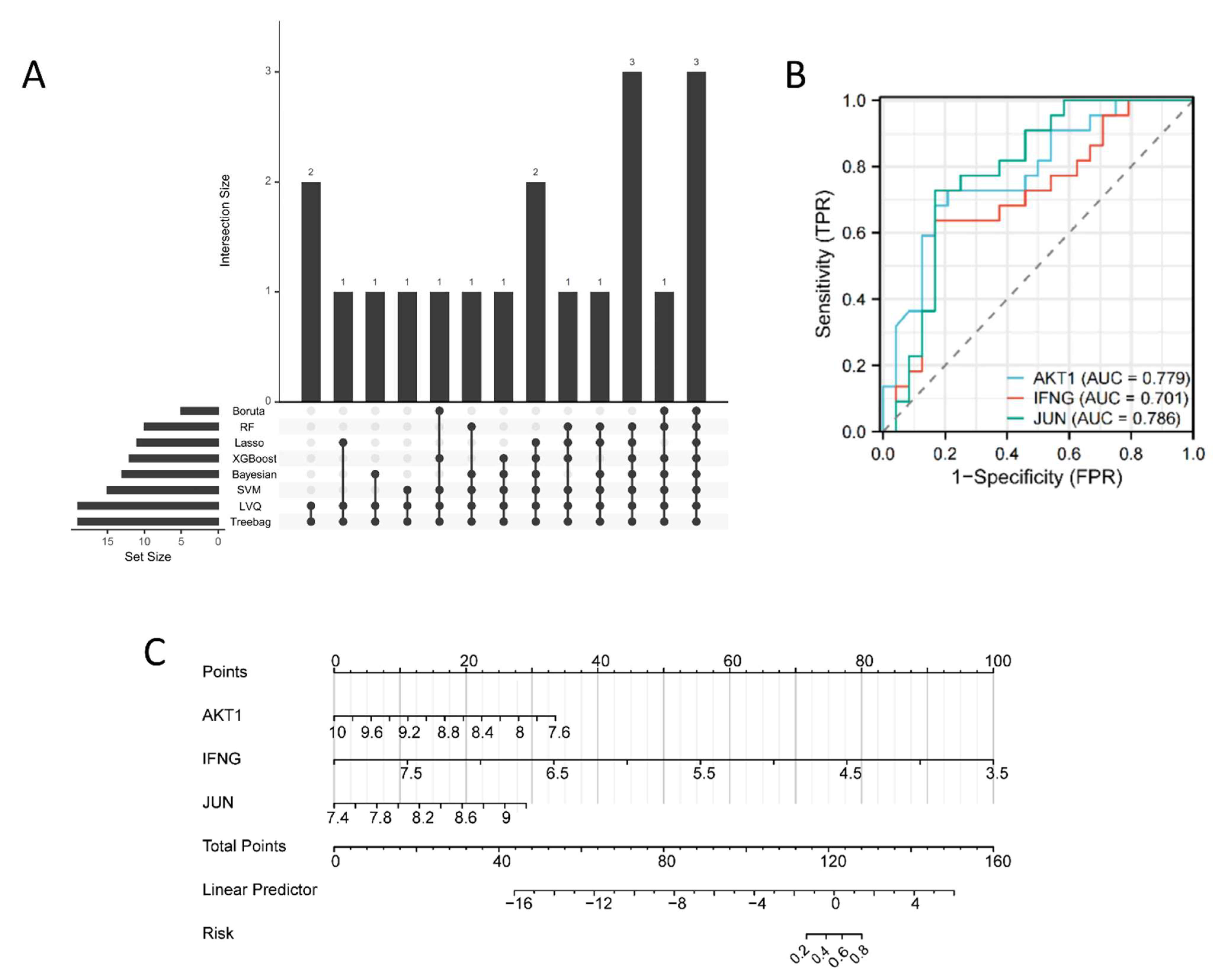
2.3. Machine Learning-Based Algorithms Identify Key Therapeutic Targets in COPD Patients
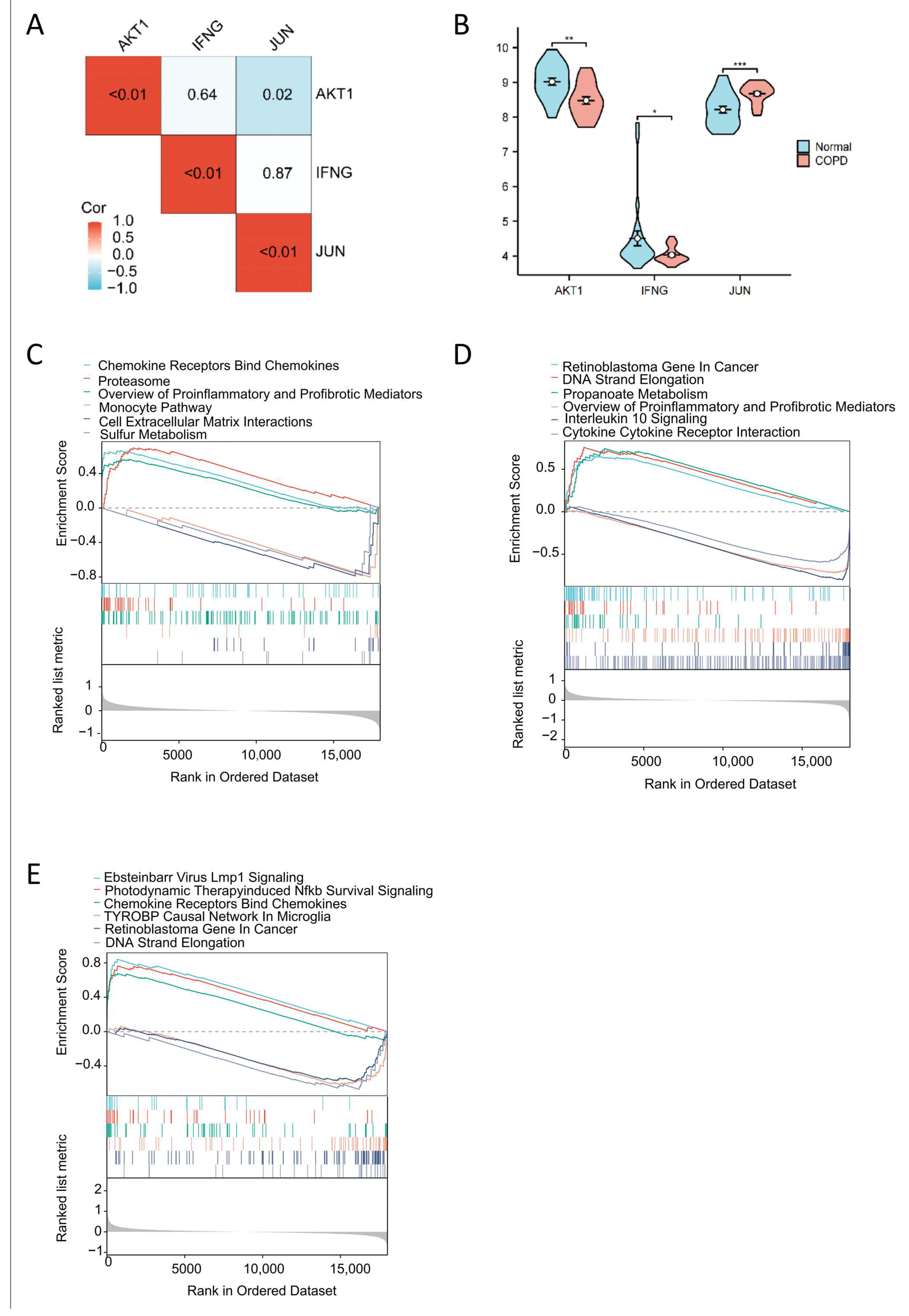
2.4. Correlation Analysis of Key Targets, COPD Expression Profiles, and Gene Set Enrichment Analysis
2.5. Immune Cell-Level Analysis
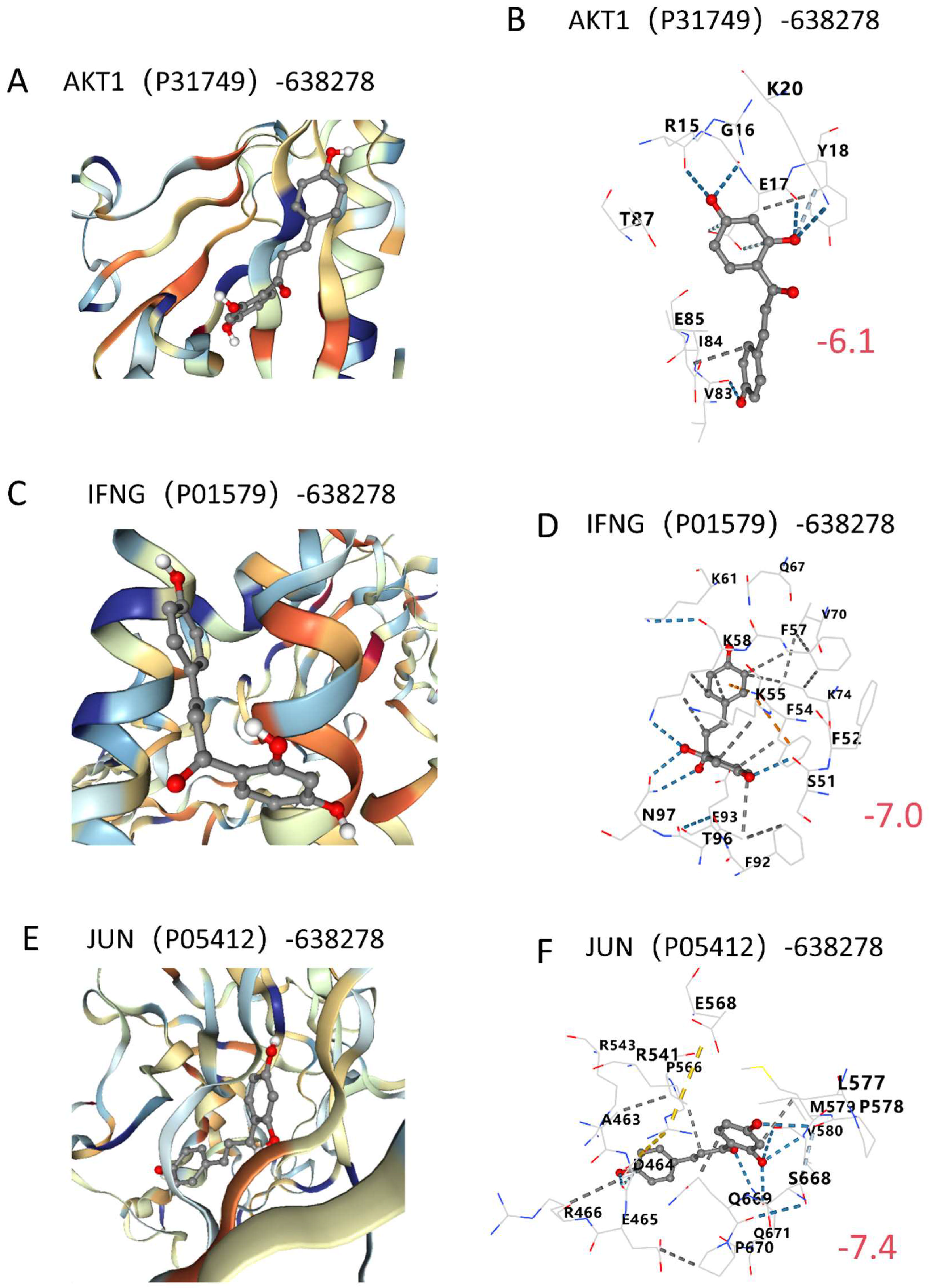
2.6. Molecular Docking Validation
3. Discussion
4. Materials and Methods
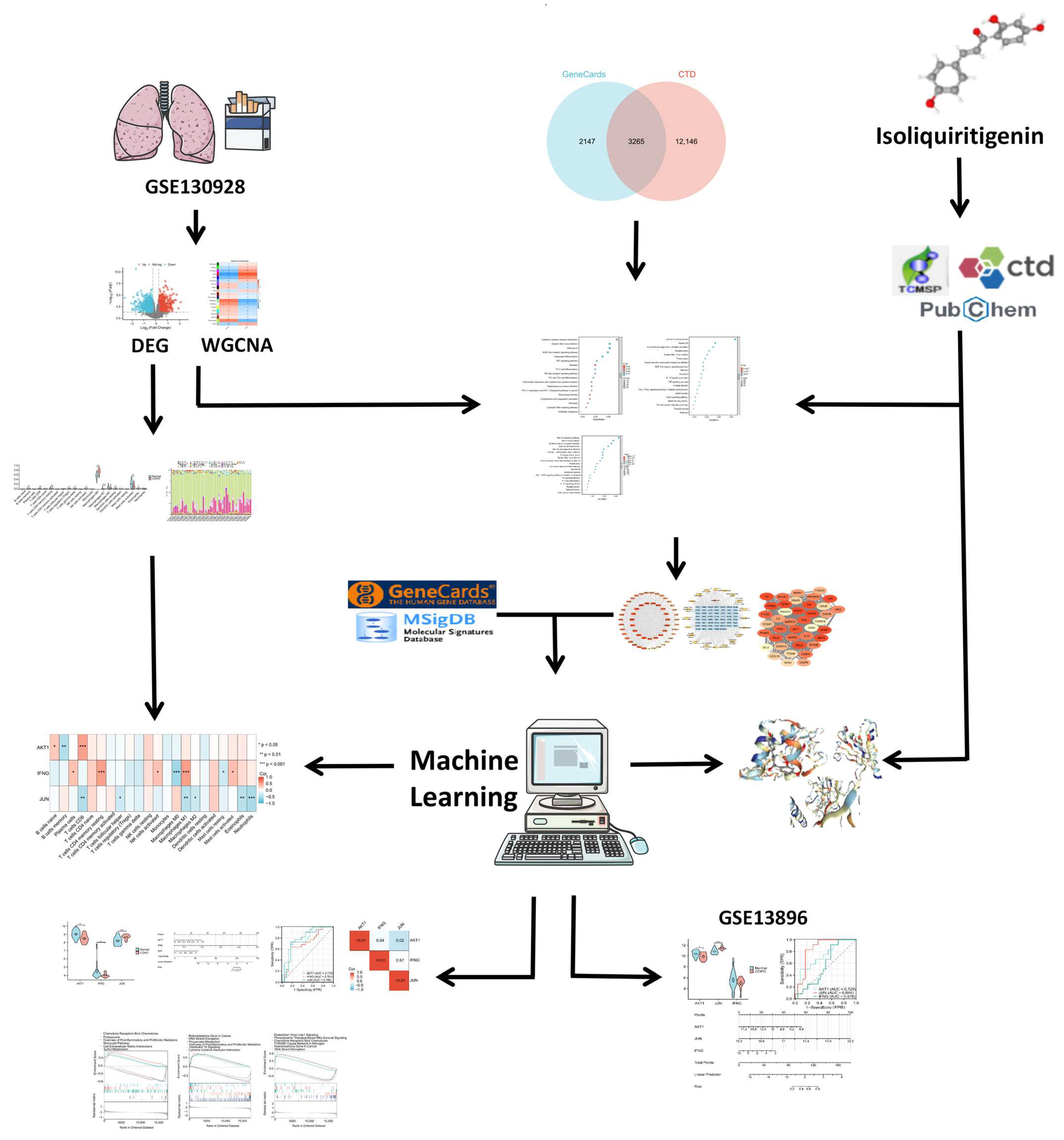
4.1. Overall Study Design
4.2. Data Description
4.3. Identification of DEGs in COPD Patients
4.4. Screening of Potential Target Genes Using WGCNA
4.5. Identification of Predicted Targets of ISO and COPD
4.6. Pathway Enrichment Analysis and Identification of Predicted Therapeutic Target Genes
4.7. Construction of PPI Networks and Small Molecule–Pathway–Target Networks
4.8. Key Goals for Screening COPD Patients Using Machine Learning Algorithms
4.9. Characterization of Key Targets for Expression, Correlation, and Gene Set Enrichment Analysis
4.10. Immune Cell Analysis
4.11. Molecular Docking Validation
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christenson, S.A.; Smith, B.M.; Bafadhel, M.; Putcha, N. Chronic obstructive pulmonary disease. Lancet 2022, 399, 2227–2242. [Google Scholar] [CrossRef] [PubMed]
- Halpin, D.M.G. Mortality of patients with COPD. Expert Rev. Respir. Med. 2024, 18, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Cadham, C.J.; Oh, H.; Han, M.K.; Mannino, D.; Cook, S.; Meza, R.; Levy, D.T.; Sánchez-Romero, L.M. The prevalence and mortality risks of PRISm and COPD in the United States from NHANES 2007–2012. Respir. Res. 2024, 25, 208. [Google Scholar] [CrossRef] [PubMed]
- Løkke, A.; Lange, P.; Lykkegaard, J.; Ibsen, R.; Andersson, M.; de Fine Licht, S.; Hilberg, O. Economic Burden of COPD by Disease Severity—A Nationwide Cohort Study in Denmark. Int. J. Chron. Obs. Pulmon Dis. 2021, 16, 603–613. [Google Scholar] [CrossRef]
- Ryan, E.M.; Sadiku, P.; Coelho, P.; Watts, E.R.; Zhang, A.; Howden, A.J.M.; Sanchez-Garcia, M.A.; Bewley, M.; Cole, J.; McHugh, B.J.; et al. NRF2 Activation Reprograms Defects in Oxidative Metabolism to Restore Macrophage Function in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2023, 207, 998–1011. [Google Scholar] [CrossRef]
- Gan, P.X.L.; Zhang, S.; Fred Wong, W.S. Targeting reprogrammed metabolism as a therapeutic approach for respiratory diseases. Biochem. Pharmacol. 2024, 228, 116187. [Google Scholar] [CrossRef]
- Aridgides, D.S.; Mellinger, D.L.; Armstrong, D.A.; Hazlett, H.F.; Dessaint, J.A.; Hampton, T.H.; Atkins, G.T.; Carroll, J.L.; Ashare, A. Functional and metabolic impairment in cigarette smoke-exposed macrophages is tied to oxidative stress. Sci. Rep. 2019, 9, 9624. [Google Scholar] [CrossRef]
- Mei, D.; Liao, W.; Gan, P.X.L.; Tran, Q.T.N.; Chan, C.C.M.Y.; Heng, C.K.M.; Wong, W.S.F. Angiotensin II type-2 receptor activation in alveolar macrophages mediates protection against cigarette smoke-induced chronic obstructive pulmonary disease. Pharmacol. Res. 2022, 184, 106469. [Google Scholar] [CrossRef]
- Wang, Z.; Li, W.; Wang, X.; Zhu, Q.; Liu, L.; Qiu, S.; Zou, L.; Liu, K.; Li, G.; Miao, H.; et al. Isoliquiritigenin induces HMOX1 and GPX4-mediated ferroptosis in gallbladder cancer cells. Chin. Med. J. 2023, 136, 2210–2220. [Google Scholar] [CrossRef]
- Yu, M.; Pan, Q.; Li, W.; Du, T.; Huang, F.; Wu, H.; He, Y.; Wu, X.; Shi, H. Isoliquiritigenin inhibits gastric cancer growth through suppressing GLUT4 mediated glucose uptake and inducing PDHK1/PGC-1α mediated energy metabolic collapse. Phytomedicine 2023, 121, 155045. [Google Scholar] [CrossRef]
- Gong, X.; Cai, J.; Zheng, W.; Huang, J.; Chen, T.; Chen, W.; Zheng, X. Isoliquiritigenin alleviates SLC7A11-mediated efferocytosis inhibition to promote wounds healing in diabetes. Biomed. Pharmacother. 2024, 180, 117578. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, Y.; Wang, Y.-Z.; Yin, P.-H.; Xu, K.; Zhang, H. The effects and mechanisms of isoliquiritigenin loaded nanoliposomes regulated AMPK/mTOR mediated glycolysis in colorectal cancer. Artif. Cells Nanomed. Biotechnol. 2020, 48, 1231–1249. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Yang, Z.; Gao, Q.; Deng, Q.; Li, L.; Chen, H. Protective Effects of Isoliquiritigenin and Licochalcone B on the Immunotoxicity of BDE-47: Antioxidant Effects Based on the Activation of the Nrf2 Pathway and Inhibition of the NF-κB Pathway. Antioxidants 2024, 13, 445. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Liu, X.; Zhang, G.; Ming, Z.; Wang, T. Isoliquiritigenin Inhibits Cigarette Smoke-Induced COPD by Attenuating Inflammation and Oxidative Stress via the Regulation of the Nrf2 and NF-κB Signaling Pathways. Front. Pharmacol. 2018, 9, 1001. [Google Scholar] [CrossRef]
- Wang, R.; Wang, M.; Ye, J.; Sun, G.; Sun, X. Mechanism overview and target mining of atherosclerosis: Endothelial cell injury in atherosclerosis is regulated by glycolysis. Int. J. Mol. Med. 2021, 47, 65–76. [Google Scholar] [CrossRef]
- Fuhrmann, D.C.; Brüne, B. miR-193a-3p increases glycolysis under hypoxia by facilitating Akt phosphorylation and PFKFB3 activation in human macrophages. Cell. Mol. Life Sci. 2022, 79, 89. [Google Scholar] [CrossRef]
- Lee, R.J.; Adappa, N.D.; Palmer, J.N. Effects of Akt Activator SC79 on Human M0 Macrophage Phagocytosis and Cytokine Production. Cells 2024, 13, 902. [Google Scholar] [CrossRef]
- Dong, Z.; Li, R.; Xu, L.; Xin, K.; Xu, Y.; Shi, H.; Sun, A.; Ge, J. Histone hyperacetylation mediates enhanced IL-1β production in LPS/IFN-γ-stimulated macrophages. Immunology 2020, 160, 183–197. [Google Scholar] [CrossRef]
- Kim, E.Y.; Ner-Gaon, H.; Varon, J.; Cullen, A.M.; Guo, J.; Choi, J.; Barragan-Bradford, D.; Higuera, A.; Pinilla-Vera, M.; Short, S.A.; et al. Post-sepsis immunosuppression depends on NKT cell regulation of mTOR/IFN-γ in NK cells. J. Clin. Investig. 2020, 130, 3238–3252. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, L.; Zhao, G.; Lin, J.; Wang, Q.; Xu, Q.; Li, C.; Hu, L.; Peng, X.; Yu, F.; et al. Indoleamine 2,3-Dioxygenase Regulates Macrophage Recruitment, Polarization and Phagocytosis in Aspergillus Fumigatus Keratitis. Invest. Ophthalmol. Vis. Sci. 2020, 61, 28. [Google Scholar] [CrossRef]
- Yu, P.-C.; Hao, C.-Y.; Fan, Y.-Z.; Liu, D.; Qiao, Y.F.; Yao, J.B.; Li, C.Z.; Yu, Y. Altered Membrane Expression and Function of CD11b Play a Role in the Immunosuppressive Effects of Morphine on Macrophages at the Nanomolar Level. Pharmaceuticals 2023, 16, 282. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, W.; Azizian, A.; Gaedcke, J.; Ohara, Y.; Cawley, H.; Hanna, N.; Ghadimi, M.; Lal, T.; Sen, S.; et al. MIF/NR3C2 axis regulates glucose metabolism reprogramming in pancreatic cancer through MAPK-ERK and AP-1 pathways. Carcinogenesis 2024, 45, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Liu, G.; Wang, X.; Lu, J.; Zhou, Y.; Chen, S.; Gao, Y.; Wang, C.; Yu, J.; Sun, Y.; et al. Transcription factor c-Jun modulates GLUT1 in glycolysis and breast cancer metastasis. BMC Cancer 2022, 22, 1283. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Duan, Y.; Yang, R.; Wang, Y.; Peng, C.; Huo, Z.; Wang, G. Foam Cell-Derived CXCL14 Muti-Functionally Promotes Atherogenesis and Is a Potent Therapeutic Target in Atherosclerosis. J. Cardiovasc. Transl. Res. 2020, 13, 215–224. [Google Scholar] [CrossRef]
- Wang, H.; Wen, C.; Chen, S.; Li, W.; Qin, Q.; He, L.; Wang, F.; Chen, J.; Ye, W.; Li, W.; et al. ROS/JNK/C-Jun Pathway is Involved in Chaetocin Induced Colorectal Cancer Cells Apoptosis and Macrophage Phagocytosis Enhancement. Front. Pharmacol. 2021, 12, 729367. [Google Scholar] [CrossRef]
- Yang, L.; Nie, H.; Du, Y.; Liu, X.; Cai, B.; Li, J. Isoliquiritigenin Exhibits Anti-Inflammatory Responses in Acute Lung Injury by Covalently Binding to the Myeloid Differentiation Protein-2 Domain. Phytother. Res. 2025, 39, 922–937. [Google Scholar] [CrossRef]
- Cui, Y.; Wu, Y.; Wang, C.; Wang, Z.; Li, Y.; Jiang, Z.; Zhao, W.; Pan, Z. Isoliquiritigenin inhibits non-small cell lung cancer progression via m6A/IGF2BP3-dependent TWIST1 mRNA stabilization. Phytomedicine 2022, 104, 154299. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, R.; Lv, M.; Li, Z.; Jin, L.; Chen, X.; Han, Y.; Shi, C.; Jiang, Y.; Jin, S. Machine-Learning Algorithm-Based Prediction of Diagnostic Gene Biomarkers Related to Immune Infiltration in Patients With Chronic Obstructive Pulmonary Disease. Front. Immunol. 2022, 13, 740513. [Google Scholar] [CrossRef]
- Kim, G.-D.; Lim, E.Y.; Shin, H.S. Macrophage Polarization and Functions in Pathogenesis of Chronic Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2024, 25, 5631. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, Z.; Peng, S.; Xu, J.; Chen, Y.; Wei, S.; Liu, X. Exosome microRNA-125a-5p derived from epithelium promotes M1 macrophage polarization by targeting IL1RN in chronic obstructive pulmonary disease. Int. Immunopharmacol. 2024, 137, 112466. [Google Scholar] [CrossRef]
- Preteroti, M.; Wilson, E.T.; Eidelman, D.H.; Baglole, C.J. Modulation of pulmonary immune function by inhaled cannabis products and consequences for lung disease. Respir. Res. 2023, 24, 95. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Wiegers, J.; Wyatt, B.; Johnson, R.J.; Sciaky, D.; Barkalow, F.; Strong, M.; Planchart, A.; Mattingly, C.J. CTD tetramers: A new online tool that computationally links curated chemicals, genes, phenotypes, and diseases to inform molecular mechanisms for environmental health. Toxicol. Sci. 2023, 195, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Dai, L.; Yuan, W.; Jiang, R.; Zhan, Z.; Zhang, L.; Xu, X.; Qian, Y.; Yang, W.; Zhang, Z. Machine learning-based integration identifies the ferroptosis hub genes in nonalcoholic steatohepatitis. Lipids Health Dis. 2024, 23, 23. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, W.; Yang, J.; Sang, Y.; Zhen, H.; Tan, C.; Huang, C.; She, J.; Liu, L.; Li, W.; et al. Enhanced machine learning models for predicting one-year mortality in individuals suffering from type A aortic dissection. J. Thorac. Cardiovasc. Surg. 2024, 169, 1191–1200. [Google Scholar] [CrossRef]
- Yan, F.; Chen, X.; Quan, X.; Wang, L.; Wei, X.; Zhu, J. Association between the stress hyperglycemia ratio and 28-day all-cause mortality in critically ill patients with sepsis: A retrospective cohort study and predictive model establishment based on machine learning. Cardiovasc. Diabetol. 2024, 23, 163. [Google Scholar] [CrossRef]
- Ning, J.; Fan, X.; Sun, K.; Wang, X.; Li, H.; Jia, K.; Ma, C. Single-cell Sequence Analysis Combined with Multiple Machine Learning to Identify Markers in Sepsis Patients: LILRA5. Inflammation 2023, 46, 1236–1254. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shao, J.; Peng, B.; Guo, Q.; Li, P.; Sun, J.; Wang, Y. Breast Tumor Diagnosis Based on Molecular Learning Vector Quantization Neural Networks. Adv. Sci. 2024, 11, e2409150. [Google Scholar] [CrossRef] [PubMed]
- O’Beirne, S.L.; Kikkers, S.A.; Oromendia, C.; Salit, J.; Rostmai, M.R.; Ballman, K.V.; Kaner, R.J.; Crystal, R.G.; Cloonan, S.M. Alveolar Macrophage Immunometabolism and Lung Function Impairment in Smoking and Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2020, 201, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Shaykhiev, R.; Krause, A.; Salit, J.; Strulovici-Barel, Y.; Harvey, B.G.; O’Connor, T.P.; Crystal, R.G. Smoking-dependent reprogramming of alveolar macrophage polarization: Implication for pathogenesis of chronic obstructive pulmonary disease. J. Immunol. 2009, 183, 2867–2883. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]


| KEGG Pathways | KEGGa Count | KEGGa p-Value | KEGGb Count | KEGGb p-Value | KEGGc Count | KEGGc p-Value | SUM. Count | AVG. p-Value |
|---|---|---|---|---|---|---|---|---|
| Influenza A | 22 | 0.001 | 94 | <0.001 | 19 | <0.001 | 135 | <0.001 |
| Osteoclast differentiation | 18 | 0.001 | 71 | <0.001 | 15 | <0.001 | 104 | <0.001 |
| NOD-like receptor signaling pathway | 21 | 0.004 | 81 | <0.001 | 23 | <0.001 | 125 | 0.001 |
| Epstein–Barr virus infection | 22 | 0.004 | 120 | <0.001 | 25 | <0.001 | 167 | 0.001 |
| TNF signaling pathway | 15 | 0.005 | 74 | <0.001 | 21 | <0.001 | 110 | 0.002 |
| Toll-like receptor signaling pathway | 14 | 0.007 | 63 | <0.001 | 18 | <0.001 | 95 | 0.0023 |
| Th1 and Th2 cell differentiation | 13 | 0.007 | 52 | <0.001 | 12 | <0.001 | 77 | 0.0023 |
| Th17 cell differentiation | 14 | 0.008 | 70 | <0.001 | 17 | <0.001 | 101 | 0.003 |
| PD-L1 expression and PD-1 checkpoint pathway in cancer | 12 | 0.013 | 53 | <0.001 | 16 | <0.001 | 81 | 0.0043 |
| Viral protein interaction with cytokine and cytokine receptor | 12 | 0.029 | 65 | <0.001 | 4 | <0.001 | 81 | 0.0397 |
| Cytosolic DNA-sensing pathway | 9 | 0.029 | 24 | <0.001 | 6 | <0.001 | 39 | 0.021 |
| Pertussis | 10 | 0.029 | 51 | <0.001 | 17 | <0.001 | 78 | 0.01 |
| Antifolate resistance | 6 | 0.029 | 17 | <0.001 | 5 | <0.001 | 28 | 0.011 |
| Rheumatoid arthritis | 11 | 0.039 | 57 | <0.001 | 6 | <0.001 | 74 | 0.015 |
| Measles | 14 | 0.046 | 77 | <0.001 | 23 | <0.001 | 114 | 0.015 |
| Chemokine signaling pathway | 17 | 0.064 | 102 | <0.001 | 14 | <0.001 | 133 | 0.022 |
| Chagas disease | 11 | 0.068 | 63 | <0.001 | 21 | <0.001 | 95 | 0.023 |
| Coronavirus disease—COVID-19 | 19 | 0.081 | 102 | <0.001 | 16 | <0.001 | 137 | 0.027 |
| Inflammatory bowel disease | 8 | 0.092 | 52 | <0.001 | 10 | <0.001 | 70 | 0.031 |
| Human T-cell leukemia virus 1 infection | 18 | 0.099 | 121 | <0.001 | 21 | <0.001 | 160 | 0.033 |
| Gene | Uniprot ID | PDB ID | Macromolecule | Center X | Center Y | Center Z | Size X | Size Y | Size Z |
|---|---|---|---|---|---|---|---|---|---|
| AKT1 | P31749 | 1H10 | AKT Serine/Threonine Kinase 1 | 23 | 19 | 22 | 22 | 22 | 22 |
| IFNG | P01579 | 1EKU | Interferon Gamma | 52 | 49 | 52 | 22 | 22 | 22 |
| JUN | P05412 | 1A02 | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | 25 | 21 | 60 | 35 | 22 | 22 |
| GEO Datasets | Normal Non-Smoking/Case | COPD/Case | Species | Method | Organization Source | Type | Platform |
|---|---|---|---|---|---|---|---|
| GSE130928 [43] | 24 | 22 | Homo sapiens | Bronchoalveolar lavage | Alveolar macrophages | Expression profiling by array | GPL570 |
| GSE13896 [44] | 24 | 12 | Homo sapiens | Bronchoalveolar lavage | Alveolar macrophages | Expression profiling by array | GPL570 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Zhang, L.; Liu, X. Bioinformatics Approach to Identifying Molecular Targets of Isoliquiritigenin Affecting Chronic Obstructive Pulmonary Disease: A Machine Learning Pharmacology Study. Int. J. Mol. Sci. 2025, 26, 3907. https://doi.org/10.3390/ijms26083907
Huang S, Zhang L, Liu X. Bioinformatics Approach to Identifying Molecular Targets of Isoliquiritigenin Affecting Chronic Obstructive Pulmonary Disease: A Machine Learning Pharmacology Study. International Journal of Molecular Sciences. 2025; 26(8):3907. https://doi.org/10.3390/ijms26083907
Chicago/Turabian StyleHuang, Sha, Lulu Zhang, and Xiaoju Liu. 2025. "Bioinformatics Approach to Identifying Molecular Targets of Isoliquiritigenin Affecting Chronic Obstructive Pulmonary Disease: A Machine Learning Pharmacology Study" International Journal of Molecular Sciences 26, no. 8: 3907. https://doi.org/10.3390/ijms26083907
APA StyleHuang, S., Zhang, L., & Liu, X. (2025). Bioinformatics Approach to Identifying Molecular Targets of Isoliquiritigenin Affecting Chronic Obstructive Pulmonary Disease: A Machine Learning Pharmacology Study. International Journal of Molecular Sciences, 26(8), 3907. https://doi.org/10.3390/ijms26083907





