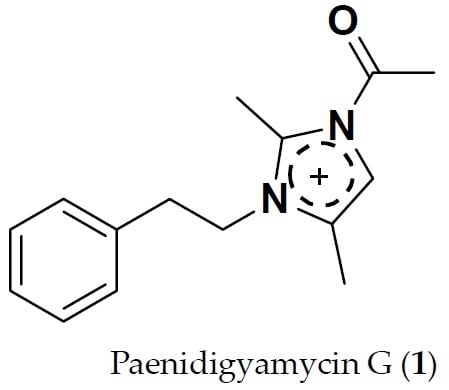
Paenidigyamycin G: 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium
Abstract
:1. Introduction
2. Results
2.1. Sediment Sample Collection Sites
2.2. Taxonomy of Strain DE2SH
2.3. Structure Determination of Paenidigyamycin G (1)
3. Experimental Section
3.1. General Experimental Procedures
3.2. Isolation and Purification of the Paenibacillus sp. Strain DE2SH
3.3. Fermentation of Paenibacillus sp. Strain DE2SH
Extraction and Purification
3.4. 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium (1)
3.5. Culture of Parasites and Mammalian Cell Lines
3.6. Analysis of Cell Viability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Vries, H.; Wagelmans, A.P.; Hasker, E.; Lumbala, C.; Lutumba, P.; De Vlas, S.J.; van de Klundert, J. Forecasting Human African Trypanosomiasis prevalences from population screening data using continuous time models. PLoS Comput. Biol. 2016, 12, e1005103. [Google Scholar] [CrossRef] [PubMed]
- Copeland, N.K.; Aronson, N.E. Leishmaniasis: Treatment updates and clinical practice guidelines review. Curr. Opin. Infect. Dis. 2015, 28, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef]
- Rollinson, D.; Knopp, S.; Levitz, S.; Stothard, J.R.; Tchuenté, L.A.T.; Garba, A.; Utzinger, J. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013, 128, 423–440. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Gaithuma, A.K.; Yamagishi, J.; Martinelli, A.; Hayashida, K.; Kawai, N.; Marsela, M.; Sugimoto, C. A single test approach for accurate and sensitive detection and taxonomic characterization of trypanosomes by comprehensive analysis of internal transcribed spacer 1 amplicons. PLoS Negl. Trop. Dis. 2019, 13, e0006842. [Google Scholar] [CrossRef]
- Mitra, A.; Mawson, A. Neglected tropical diseases: Epidemiology and global burden. Trop. Med. Infect. Dis. 2017, 2, 36. [Google Scholar] [CrossRef]
- Hotez, P.J.; Kamath, A. Neglected tropical diseases in sub-Saharan Africa: Review of their prevalence, distribution, and disease burden. PLoS Negl. Trop. Dis. 2009, 3, e412. [Google Scholar] [CrossRef]
- Hotez, P.J.; Pecoul, B.; Rijal, S.; Boehme, C.; Aksoy, S.; Malecela, M.; Reeder, J.C. Eliminating the neglected tropical diseases: Translational science and new technologies. PLoS Negl. Trop. Dis. 2016, 10, e0003895. [Google Scholar] [CrossRef]
- Pink, R.; Hudson, A.; Mouriès, M.A.; Bendig, M. Opportunities and challenges in antiparasitic drug discovery. Nat. Rev. Drug Discov. 2005, 4, 727–740. [Google Scholar] [CrossRef]
- Cheuka, P.; Mayoka, G.; Mutai, P.; Chibale, K. The role of natural products in drug discovery and development against neglected tropical diseases. Molecules 2017, 22, 58. [Google Scholar] [CrossRef] [PubMed]
- Addisu, A.; Adriaensen, W.; Balew, A.; Asfaw, M.; Diro, E.; Djirmay, A.G.; Adugna, A.H. Neglected tropical diseases and the sustainable development goals: An urgent call for action from the front line. BMJ Glob Health. 2019, 4, e001334. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.G.; Kumar, V.; Kaur, K. Imidazole containing natural products as antimicrobial agents: A review. Nat. Prod. J. 2014, 4, 73–81. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Kharb, R.; Kumar, S.; Sharma, C.P.; Pathak, P.D. Imidazole derivatives as potential therapeutic agents. Curr. Pharm. Des. 2016, 22, 3265–3301. [Google Scholar] [CrossRef]
- Luca, L.D. Naturally occurring and synthetic imidazoles: Their chemistry and their biological activities. Curr. Med. Chem. 2006, 13, 1–23. [Google Scholar]
- Liu, C.; Shi, C.; Mao, F.; Xu, Y.; Liu, J.; Wei, B.; Li, J. Discovery of new imidazole derivatives containing the 2,4-dienone motif with broad-spectrum antifungal and antibacterial activity. Molecules 2014, 19, 15653–15672. [Google Scholar] [CrossRef]
- Li, X.; Zhan, P.; Clercq, D.E.; Liu, X. The HIV-1 non-nucleoside reverse transcriptase inhibitors (Part V*): Capravirine and its analogues. Curr. Med. Chem. 2012, 19, 6138–6149. [Google Scholar]
- Zhang, W.; Shen, X.; Bergman, U.; Wang, Y.; Chen, Y.; Huang, M.; Deng, L. Drug utilisation 90%(DU90%) profiles of antibiotics in five Chinese children’s hospitals (2002–2006). Int. J. Antimicrob. Agents 2008, 32, 250–255. [Google Scholar] [CrossRef]
- Rotta, I.; Ziegelmann, P.K.; Otuki, M.F.; Riveros, B.S.; Bernardo, N.L.; Correr, C.J. Efficacy of topical antifungals in the treatment of dermatophytosis: A mixed-treatment comparison meta-analysis involving 14 treatments. JAMA Dermatol. 2013, 149, 341–349. [Google Scholar] [CrossRef]
- Gaba, M.; Mohan, C. Development of drugs based on imidazole and benzimidazole bioactive heterocycles: Recent advances and future directions. Med. Chem. Res. 2016, 25, 173–210. [Google Scholar] [CrossRef]
- Abdellatif, K.R.A.; Fadaly, W.A.A. New 1,2-diaryl-4-substituted-benzylidene-5-4H-imidazolone derivatives: Design, synthesis and biological evaluation as potential anti-inflammatory and analgesic agents. Bioorg. Chem. 2017, 72, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Acar, M.F.; Sari, S.; Dalkara, S. Synthesis, in vivo anticonvulsant testing, and molecular modeling studies of new nafimidone derivatives. Drug Dev. Res. 2019, 80, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Watts, A.E.; Maruyoshi, K.; Hughes, C.E.; Brown, S.P.; Harris, K.D. Combining the advantages of powder X-ray diffraction and NMR crystallography in structure determination of the pharmaceutical material cimetidine hydrochloride. Cryst. Growth Des. 2016, 16, 1798–1804. [Google Scholar] [CrossRef]
- Osei, E.; Kwain, S.; Tetevi, M.G.; Anang, K.A.; Owusu, K.B.-A.; Camas, M.; Camas, A.S.; Ohashi, M.; Alexandru-Crivac, C.-N.; Deng, H.; et al. Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH. Mar. Drugs 2019, 17, 9. [Google Scholar] [CrossRef]
- Kwain, S.; Tetevi, G.M.; Mensah, T.; Camas, A.S.; Camas, M.; Dofuor, A.K.; Azerigyik, F.A.; Deng, H.; Jaspars, M.; Kyeremeh, K. Digyaindoleacid A: 2-(1-(4-Hydroxyphenyl)-3-oxobut-1-en-2-yloxy) -3-(1H-indol-3-yl) propanoic Acid, a Novel Indole Alkaloid. Molbank 2019, 2019, M1080. [Google Scholar] [CrossRef]
- Yabu, Y.; Minagawa, N.; Kita, K.; Nagai, K.; Honma, M.; Sakajo, S.; Koide, T.; Ohta, N.; Yoshimoto, A. Oral and intraperitoneal treatment of Trypanosoma brucei brucei with a combination of ascofuranone and glycerol in mice. Parasitol. Int. 1998, 47, 131–137. [Google Scholar] [CrossRef]




| # | δC mult | δH mult (J Hz) | 1H-1H COSY | HMBC | TOCSY |
|---|---|---|---|---|---|
| 1-N | |||||
| 2 | 137.3, C | ||||
| 3-N | |||||
| 4 | 116.5, C | ||||
| 5 | 122.3, CH | 6.96, s | 6 | C-2, C-4, C-6 | |
| 6 | 9.7, CH3 | 1.99, s | 5 | C-4, C-5, C-2 | |
| 7 | 8.2, CH3 | 1.90, s | C-4, C-2 | ||
| 8 | 47.0, CH2 | 4.48, t (7.2) | 9 | C-2, C-9 | 9 |
| 9 | 36.9, CH2 | 2.92, t (7.2) | 2’, 8 | C-2’, C-1’, C-8 | 8 |
| 10 | 187.5, C | ||||
| 11 | 25.4, CH3 | 2.39, s | C-10 | ||
| 1’ | 138.7, C | ||||
| 2’ | 128.7, CH | 7.09, m | 3’, 9 | C-4’, C-6’, C-9 | 3’ |
| 3’ | 128.0, CH | 7.25, m | 2’ | C-5’, C-1’ | 2’, 6’ |
| 4’ | 126.0, CH | 7.20, m | 3’, 2’, 6’ | ||
| 5’ | 128.0, CH | 7.25, m | 2’ | C-3’, C-1’ | 2’, 6’ |
| 6’ | 128.7, CH | 7.09, m | 3’ | C-4’, C-2’, C-9 | 3’ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tetevi, G.M.; Kwain, S.; Mensah, T.; Camas, A.S.; Camas, M.; Dofuor, A.K.; Azerigyik, F.A.; Oluwabusola, E.; Deng, H.; Jaspars, M.; et al. Paenidigyamycin G: 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium. Molbank 2019, 2019, M1094. https://doi.org/10.3390/M1094
Tetevi GM, Kwain S, Mensah T, Camas AS, Camas M, Dofuor AK, Azerigyik FA, Oluwabusola E, Deng H, Jaspars M, et al. Paenidigyamycin G: 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium. Molbank. 2019; 2019(4):M1094. https://doi.org/10.3390/M1094
Chicago/Turabian StyleTetevi, Gilbert Mawuli, Samuel Kwain, Thomas Mensah, Anil Sazak Camas, Mustafa Camas, Aboagye Kwarteng Dofuor, Faustus Akankperiwen Azerigyik, Emmanuel Oluwabusola, Hai Deng, Marcel Jaspars, and et al. 2019. "Paenidigyamycin G: 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium" Molbank 2019, no. 4: M1094. https://doi.org/10.3390/M1094
APA StyleTetevi, G. M., Kwain, S., Mensah, T., Camas, A. S., Camas, M., Dofuor, A. K., Azerigyik, F. A., Oluwabusola, E., Deng, H., Jaspars, M., & Kyeremeh, K. (2019). Paenidigyamycin G: 1-Acetyl-2,4-dimethyl-3-phenethyl-1H-imidazol-3-ium. Molbank, 2019(4), M1094. https://doi.org/10.3390/M1094