An Apta-Biosensor for Colon Cancer Diagnostics
Abstract
:1. Introduction

2. Aptasensor Design
2.1. Aptamer Synthesis


2.2. Recognition Element
- Creation of thiol group by coating 11-MUA on electrode.
- Activation of COOH group in 11-MUA with ethyl(dimethylaminopropyl)carbodiimide (EDC)/N-hydroxysuccinimide (NHS).
- Binding with the NH2 at the 3′ end of the aptamer.
- Coating with bovine serum albumin (BSA) to prevent non-specific binding.

3. Materials and Methods
3.1. Apta-Sensor Development
3.2. Cell Culture
3.3. Flow Cytometry & Fluorescent Microscopy
3.4. Electrochemical Experiments
4. Results
4.1. Fluorescence Microscopy

4.2. Flow Cytometry

| %Gated (R1 Region) | Without Aptamer | Aptamer Attached | |
|---|---|---|---|
| Cell Line | |||
| HEp-2 | 0.2 | 0.2 | |
| HCT 116 | 0.43 | 60.19 | |
| HT 29 | 0.40 | 29.62 | |
4.3. Electrochemical Experiments
4.3.1. Functionalization of Electrodes

4.3.2. Elctrochemical Biosensor

5. Discussion
5.1. Self-Assembly of Monolayers on Electrodes
5.2. Colon Cancer Cell Detection

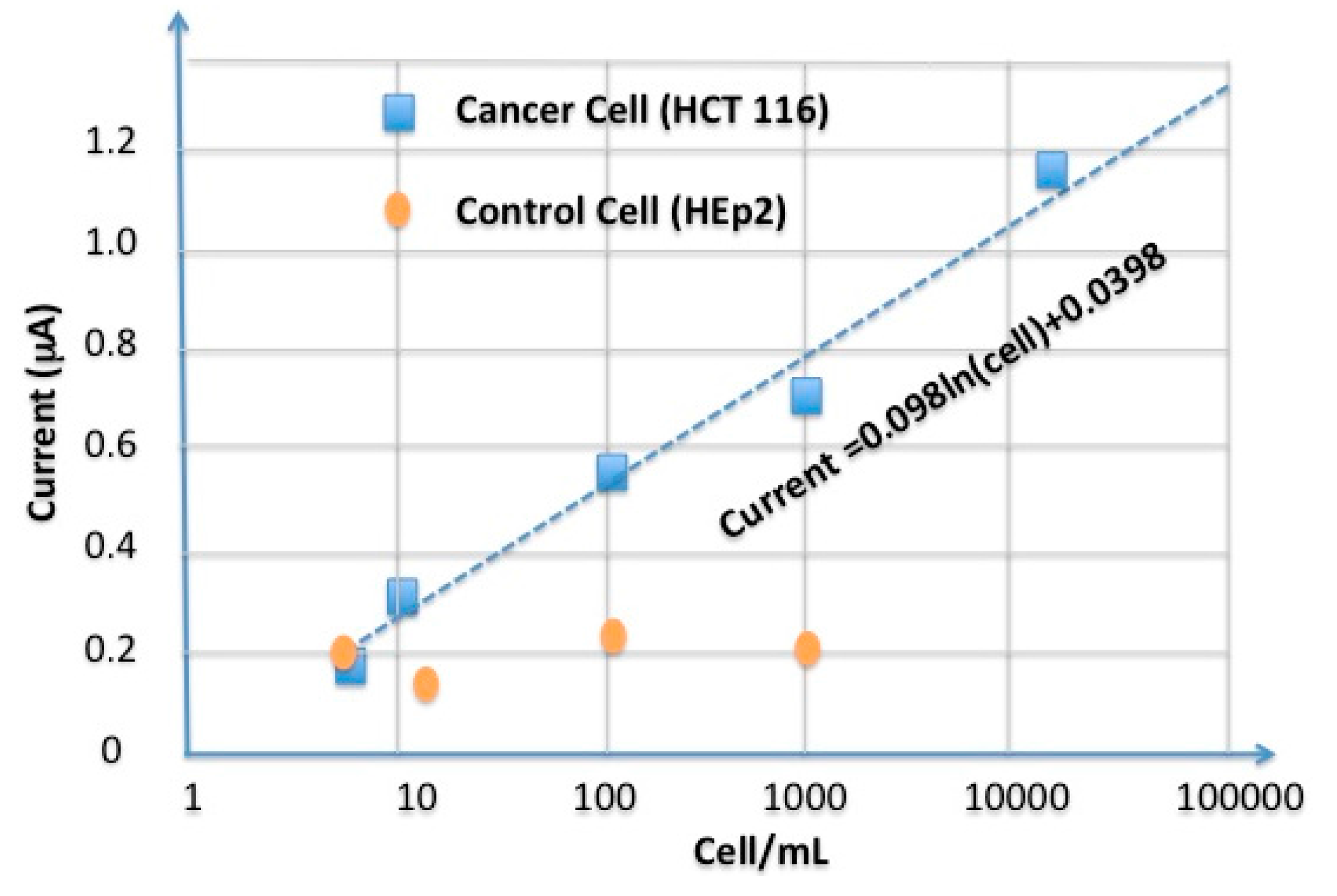
5.3. Sensitivity of the Aptasensor
5.4. Selectivity of the Aptasensor
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fast Facts on Colorectal Cancer (CRC). Available online: http://coloncancercanada.ca/fast-facts-on-colorectal-cancer-crc/ (accessed on 1 May 2015).
- 2014 Canadian Cancer Statistics. Available online: http://www.colorectal-cancer.ca/en/just-the-facts/colorectal/ (accessed on 1 May 2015).
- Dassie, J.P.; Hernandez, L.I.; Thomas, G.S.; Long, M.E.; Rockey, W.M.; Howell, C.A.; Chen, Y.; Hernandez, F.J.; Liu, X.Y.; Wilson, M.E.; et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol. Ther. 2014, 22, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Ye, D.; Zhao, Q.; Luo, J.; Zhang, S.; Kong, J. A novel apta-sensor based on MUC-1 conjugated CNSs for ultrasensitive detection of tumor cells. Analyst 2014, 139, 4917–4923. [Google Scholar] [CrossRef] [PubMed]
- Pourhoseingholi, M.A.; Zali, M.R. Colorectal cancer screening: Time for action in Iran. World J. Gastrointest. Oncol. 2012, 4, 82–83. [Google Scholar] [CrossRef] [PubMed]
- Chenga, A.K.H.; Sena, D.; Yua, H.Z. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 2009, 77, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mascinis, M.; Tombelli, S. Biosensors for biomarkers in medical diagnostics. Biomarkers 2008, 13, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.M.; Donovan, M.J.; Tan, W. Using Aptamers for Cancer Biomarker Discovery. J. Nucleic Acids 2013, 2013, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hammarstro, S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Obernik, B. CEA adhesion molecules: Multifunctional proteins with signal-regulatory properties. Curr. Opin. Cell Biol. 1997, 9, 616–626. [Google Scholar] [CrossRef]
- Sadava, D.E. Cell Biology-Organelle Structure and Function, 1st ed.; Jones and Bartlett: Charlestown, MA, USA, 1993; Chapter 1; p. 22. [Google Scholar]
- Sefah, K.; Meng, L.; Lopez-Colon, D.; Jimenez, E.; Liu, C.H.; Tan, W. DNA Aptamers as Molecular Probes for Colorectal Cancer Study. PLoS ONE 2010, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Reth, M. Matching cellular dimensions with molecular sizes. Nat. Immunol. 2013, 14, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Medley, C.D.; Smith, J.E.; Tang, Z.; Wu, Y.; Bamrungsap, S.; Tan, W. Gold Nanoparticle-Based Colorimetric Assay for the Direct Detection of Cancerous Cells. Anal. Chem. 2008, 80, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C. Methods developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.C.; Huang, C.C. Aptamer-Functionalized Nano-Biosensors. Sensors 2009, 9, 10356–10388. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tuleouva, N.; Ramanculov, E.; Revzin, A. Aptamer-Based Electrochemical Biosensor for Interferon Gamma Detection. Anal. Chem. 2010, 82, 8131–8136. [Google Scholar] [CrossRef] [PubMed]
- Bang, G.S.; Cho, S.; Kim, B.G. A novel electrochemical detection method for aptamer biosensors. Biosens. Bioelectron. 2005, 21, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Singh, J.; Singh, A.; Srivastava, A.; Goyal, R.N.; Shim, Y.B. Gold Nanoparticles and Nanocomposites in Clinical Diagnostics Using Electrochemical Methods. J. Nanopart. 2013, 2013. [Google Scholar] [CrossRef]
- Ocaña, C.; Pacios, M.; del Valle, M. A Reusable Impedimetric Aptasensor for Detection of Thrombin Employing a Graphite-Epoxy Composite Electrode. Sensors 2012, 12, 3037–3048. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; Soper, S.A.; Spivak, D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008, 390, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; McCarley, R.L.; Soper, S.A.; Spivak, D.A. Effect of Linker Structure on Surface Density of Aptamer Monolayers and Their Corresponding Protein Binding Efficiency. Anal. Chem. 2008, 80, 9630–9634. [Google Scholar] [CrossRef] [PubMed]
- KineFold Simulation Results. Available online: http://kinefold.curie.fr/cgi-bin/neorequest.pl?batch=0&sim=2-&base=KCHA10a-49060 (accessed on 30 April 2015).
- Hermanson, G.T. Bioconjugate Techniques, 2nd ed.; Academic Press, Inc. of Elsevier: New York, NY, USA, 2008. [Google Scholar]
- Freshney, R.I. Culture of Animal Cells a Manual of Basic Technique, 5th ed.; Wiley: New York City, NJ, USA, 2005; pp. 199–215. [Google Scholar]
- Sheng, W.; Chen, T.; Kamath, R.; Xiong, X.; Tan, W.; Fan, Z.H. Aptamer-Enabled Efficient Isolation of Cancer Cells from Whole Blood Using a Micro fluidic Device. Anal. Chem. 2012, 84, 4199–4206. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raji, M.A.; Amoabediny, G.; Tajik, P.; Hosseini, M.; Ghafar-Zadeh, E. An Apta-Biosensor for Colon Cancer Diagnostics. Sensors 2015, 15, 22291-22303. https://doi.org/10.3390/s150922291
Raji MA, Amoabediny G, Tajik P, Hosseini M, Ghafar-Zadeh E. An Apta-Biosensor for Colon Cancer Diagnostics. Sensors. 2015; 15(9):22291-22303. https://doi.org/10.3390/s150922291
Chicago/Turabian StyleRaji, Mojgan Ahmadzadeh, Ghasem Amoabediny, Parviz Tajik, Morteza Hosseini, and Ebrahim Ghafar-Zadeh. 2015. "An Apta-Biosensor for Colon Cancer Diagnostics" Sensors 15, no. 9: 22291-22303. https://doi.org/10.3390/s150922291






