Label-Free Biosensor Imaging on Photonic Crystal Surfaces
Abstract
:1. Introduction



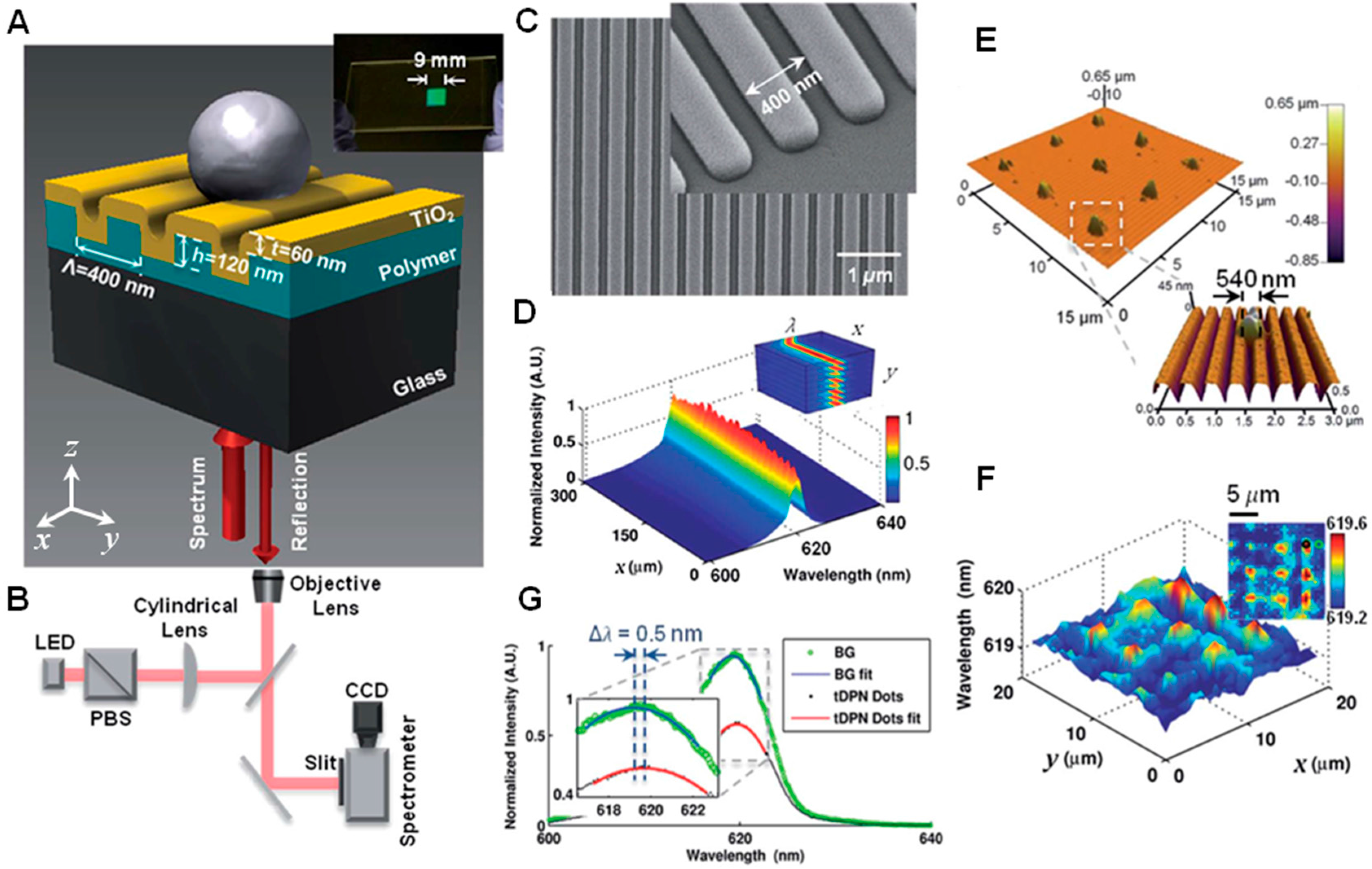
2. Principles of Modern PCEM
2.1. Photonic Crystal Surface Biosensor
2.2. PCEM Imaging Modality and Operating Principle
3. History of PCEM Development
3.1. Instrument 1—Biomolecular Interaction Detection (BIND) Scanner
3.2. Instrument 2—Transmission Acquisition Mode with Upright Microscopy and Laser Source
3.3. Instrument 3—Reflection Acquisition Mode with Inverted Microscopy and LED Source
4. Applications of PCEM
4.1. PCEM for Label-Free Live Cell Imaging

4.2. PCEM for Imaging of Nanoparticle and Protein-Protein Binding


4.3. Combination of PCEM and PCEF for Label-Free/Fluorescence-Labeled Imaging Simultaneously
5. Summary
Acknowledgments
Conflicts of Interest
References
- Hessel, A.; Oliner, A.A. A new theory of wood’s anomalies on optical gratings. Appl. Opt. 1965, 4, 1275–1297. [Google Scholar] [CrossRef]
- Mashev, L.; Popov, E. Diffraction efficiency anomalies of multicoated dielectric gratings. Opt. Commun. 1984, 51, 131–136. [Google Scholar] [CrossRef]
- Popov, E.; Mashev, L.; Maystre, D. Theoretical study of the anomalies of coated dielectric gratings. Opt. Acta 1986, 33, 607–619. [Google Scholar] [CrossRef]
- Yablonovitch, E. Inhibited spontaneous emission in solid-state physics and electronics. Phys. Rev. Lett. 1987, 58, 2059–2062. [Google Scholar] [CrossRef] [PubMed]
- John, S. Strong localization of photons in certain disordered dielectric superlattices. Phys. Rev. Lett. 1987, 58, 2486–2489. [Google Scholar] [CrossRef] [PubMed]
- Joannopoulos, J.D.; Villeneuve, P.R.; Fan, S. Photonic crystals: Putting a new twist on light. Nature 1997, 386, 143–149. [Google Scholar] [CrossRef]
- Fan, S.H.; Joannopoulos, J.D. Analysis of guided resonances in photonic crystal slabs. Phys. Rev. B 2002, 65, 235112. [Google Scholar] [CrossRef]
- Joannopoulos, J.D.; Johnson, S.G.; Winn, J.N.; Meade, R.D. Photonic Crystals: Molding the Flow of Light, 2nd ed.; Princeton University Press: Princeton, NJ, USA, 2008. [Google Scholar]
- Magnusson, R.; Wang, S.S. New principle for optical filters. Appl. Phys. Lett. 1992, 61, 1022–1024. [Google Scholar] [CrossRef]
- Kikuta, H.; Maegawa, N.; Mizutani, A.; Iwata, K.; Toyota, H. Refractive index sensor with a guided-mode resonant grating filter. Proc. SPIE 2001, 4416, 219–222. [Google Scholar]
- Villa, F.; Regalado, L.E.; Ramos-Mendieta, F.; Gaspar-Armenta, J.; Lopez-Rios, T. Photonic crystal sensor based on surface waves for thin-film characterization. Opt. Lett. 2002, 27, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.T.; Li, P.; Schulz, S.; Lin, B.; Baird, C.; Gerstenmaier, J.; Genick, C.; Wang, F.; Fine, E.; Laing, L. Label-free assays on the bind system. J. Biomol. Screen. 2004, 9, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Ferrie, A.M.; Fontaine, N.H.; Mauro, J.; Balakrishnan, J. Resonant waveguide grating biosensor for living cell sensing. Biophys. J. 2006, 91, 1925–1940. [Google Scholar] [CrossRef] [PubMed]
- Skivesen, N.; Tetu, A.; Kristensen, M.; Kjems, J.; Frandsen, L.H.; Borel, P.I. Photonic-crystal waveguide biosensor. Opt. Expr. 2007, 15, 3169–3176. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. Photonic crystal surface waves for optical biosensors. Anal. Chem. 2007, 79, 4729–4735. [Google Scholar] [CrossRef] [PubMed]
- Nazirizadeh, Y.; Geyer, U.; Lemmer, U.; Gerken, M. Spatially resolved optical characterization of photonic crystal slabs using direct evaluation of photonic modes. In Proceedings of the IEEE International Conference on Optical MEMs and Nanophotonics, Freiburg, Gremany, 11 August 2008; pp. 112–113.
- Guo, Y.B.; Divin, C.; Myc, A.; Terry, F.L.; Baker, J.R.; Norris, T.B.; Ye, J.Y. Sensitive molecular binding assay using a photonic crystal structure in total internal reflection. Opt. Expr. 2008, 16, 11741–11749. [Google Scholar] [CrossRef]
- Fang, Y.; Frutos, A.G.; Verklereen, R. Label-free cell-based assays for gpcr screening. Comb. Chem. High Throughput Screen. 2008, 11, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Konopsky, V.N.; Alieva, E.V. Optical biosensors based on photonic crystal surface waves. Methods Mol. Biol. 2009, 503, 49–64. [Google Scholar] [PubMed]
- Cunningham, B.T. Photonic crystal surfaces as a general purpose platform for label-free and fluorescent assays. JALA Charlottesv Va 2010, 15, 120–135. [Google Scholar] [CrossRef] [PubMed]
- El Beheiry, M.; Liu, V.; Fan, S.; Levi, O. Sensitivity enhancement in photonic crystal slab biosensors. Opt. Expr. 2010, 18, 22702–22714. [Google Scholar] [CrossRef] [PubMed]
- Nazirizadeh, Y.; Bog, U.; Sekula, S.; Mappes, T.; Lemmer, U.; Gerken, M. Low-cost label-free biosensors using photonic crystals embedded between crossed polarizers. Opt. Expr. 2010, 18, 19120–19128. [Google Scholar] [CrossRef] [PubMed]
- Jamois, C.; Li, C.; Gerelli, E.; Orobtchouk, R.; Benyattou, T.; Belarouci, A.; Chevolot, Y.; Monnier, V.; Souteyrand, E. New Concepts of Integrated Photonic Biosensors Based on Porous Silicon. In Biosensors-Emerging Materials and Applications; Serra, P.A., Ed.; InTech: Rijeka, Croatia, 2011. [Google Scholar]
- Magnusson, R.; Wawro, D.; Zimmerman, S.; Ding, Y. Resonant photonic biosensors with polarization-based multiparametric discrimination in each channel. Sensors 2011, 11, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Nazirizadeh, Y.; Becker, T.; Reverey, J.; Selhuber-Unkel, C.; Rapoport, D.H.; Lemmer, U.; Gerken, M. Photonic crystal slabs for surface contrast enhancement in microscopy of transparent objects. Opt. Expr. 2012, 20, 14451–14459. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Fauchet, P.M.; Miller, B.L. 1-d and 2-d photonic crystals as optical methods for amplifying biomolecular recognition. Anal.Chem. 2012, 84, 8900–8908. [Google Scholar] [CrossRef] [PubMed]
- Threm, D.; Nazirizadeh, Y.; Gerken, M. Photonic crystal biosensors towards on-chip integration. J. Biophotonics 2012, 5, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, J.; Diaz-Rubio, A.; Torrent, D.; Cervera, F.; Kirleis, M.A.; Pique, A.; Sanchez-Dehesa, J. Radial photonic crystal for detection of frequency and position of radiation sources. Sci. Rep. 2012, 2, 558. [Google Scholar] [CrossRef] [PubMed]
- Grepstad, J.O.; Kaspar, P.; Solgaard, O.; Johansen, I.R.; Sudbo, A.S. Photonic-crystal membranes for optical detection of single nano-particles, designed for biosensor application. Opt. Expr. 2012, 20, 7954–7956. [Google Scholar] [CrossRef] [PubMed]
- Troia, B.; Paolicelli, A.; Leonardis, F.D.; Passaro, V.M.N. Photonic crystals for optical sensing: A review. In Advances in Photonic Crystals; Passaro, V.M.N., Ed.; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Lin, B.; Qiu, J.; Gerstenmeier, J.; Li, P.; Pien, H.; Pepper, J.; Cunningham, B. A label-free optical technique for detecting small molecule interactions. Biosens. Bioelectron. 2002, 17, 827–834. [Google Scholar] [CrossRef]
- Chan, L.L.; Cunningham, B.T.; Li, P.Y.; Puff, D. A self-referencing method for microplate label-free photonic-crystal biosensors. IEEE Sens. J. 2006, 6, 1551–1556. [Google Scholar] [CrossRef]
- Chan, L.L.; Lidstone, E.A.; Finch, K.E.; Heeres, J.T.; Hergenrother, P.J.; Cunningham, B.T. A method for identifying small molecule aggregators using photonic crystal biosensor microplates. J. Assoc. Lab. Autom. 2009, 14, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Lu, M.; George, S.; Flood, T.A.; Wagner, C.; Zheng, J.; Pokhriyal, A.; Eden, J.G.; Hergenrother, P.J.; Cunningham, B.T. External cavity laser biosensor. Lab Chip 2013, 13, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Peh, J.; Hergenrother, P.J.; Cunningham, B.T. Detection of protein-small molecule binding using a self-referencing external cavity laser biosensor. J. Am. Chem. Soc. 2014, 136, 5840–5843. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, H.; Lidstone, E.A.; Jahangir, M.; Inci, F.; Hanhauser, E.; Henrich, T.J.; Kuritzkes, D.R.; Cunningham, B.T.; Demirci, U. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci. Rep. 2014, 4, 4116. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Block, I.D.; Jones, S.I.; Mathias, P.C.; Chaudhery, V.; Vuttipittayamongkol, P.; Wu, H.Y.; Vodkin, L.O.; Cunningham, B.T. Label-free prehybridization DNA microarray imaging using photonic crystals for quantitative spot quality analysis. Anal. Chem. 2010, 82, 8551–8557. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Li, P.; Cunningham, B.T. A label-free biosensor-based cell attachment assay for characterization of cell surface molecules. Sens. Actuators B Chem. 2006, 114, 559–564. [Google Scholar] [CrossRef]
- Chan, L.L.; Gosangari, S.L.; Watkin, K.L.; Cunningham, B.T. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis 2007, 12, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.L.; Gosangari, S.L.; Watkin, K.L.; Cunningham, B.T. Label-free imaging of cancer cells using photonic crystal biosensors and application to cytotoxicity screening of a natural compound library. Sens. Actuators B Chem. 2008, 132, 418–425. [Google Scholar] [CrossRef]
- George, S.; Bhalerao, S.V.; Lidstone, E.A.; Ahmad, I.S.; Abbasi, A.; Cunningham, B.T.; Watkin, K.L. Cytotoxicity screening of bangladeshi medicinal plant extracts on pancreatic cancer cells. Complement. Altern. Med. 2010, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Cunningham, B.T. Label-free cell-based assays using photonic crystal optical biosensors. Analyst 2011, 136, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.L.; George, S.; Ahmad, I.; Gosangari, S.L.; Abbasi, A.; Cunningham, B.T.; Watkin, K.L. Cytotoxicity effects of amoorarohituka and chittagonga on breast and pancreatic cancer cells. Complement. Altern. Med. 2011, 10, 1–8. [Google Scholar]
- Lidstone, E.A.; Chaudhery, V.; Kohl, A.; Chan, V.; Wolf-Jensen, T.; Schook, L.B.; Bashir, R.; Cunningham, B.T. Label-free imaging of cell attachment with photonic crystal enhanced microscopy. Analyst 2011, 136, 3608–3615. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Long, K.D.; Lu, M.; Chaudhery, V.; Yu, H.; Choi, J.S.; Polans, J.; Zhuo, Y.; Harley, B.A.C.; Cunningham, B.T. Photonic crystal enhanced microscopy for imaging of live cell adhesion. Analyst 2013, 138, 5886–5894. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.T.; Qiu, J.; Li, P.; Pepper, J.; Hugh, B. A plastic colorimetric resonant optical biosensor for multiparallel detection of label-free biochemical interactions. Sens. Actuators B Chem. 2002, 85, 219–226. [Google Scholar] [CrossRef]
- Li, P.; Lin, B.; Gerstenmaier, J.; Cunningham, B.T. A new method for label-free imaging of biomolecular interactions. Sens. Actuators B Chem. 2004, 99, 6–13. [Google Scholar] [CrossRef]
- Zhuo, Y.; Hu, H.; Chen, W.L.; Lu, M.; Tian, L.M.; Yu, H.J.; Long, K.D.; Chow, E.; King, W.P.; Singamaneni, S.; et al. Single nanoparticle detection using photonic crystal enhanced microscopy. Analyst 2014, 139, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.; Qiu, J.; Li, P.; Lin, B. Enhancing the surface sensitivity of colorimetric resonant optical biosensors. Sens. Actuators B Chem. 2002, 87, 365–370. [Google Scholar] [CrossRef]
- Cunningham, B.T.; Laing, L. Microplate-based, label-free detection of biomolecular interactions: Applications in proteomics. Expert Rev. Proteom. 2006, 3, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.T.; Li, P.; Lin, B.; Pepper, J. Colorimetric resonant reflection as a direct biochemical assay technique. Sens. Actuators B Chem. 2002, 81, 316–328. [Google Scholar] [CrossRef]
- Yeh, P.; Yariv, A.; Cho, A.Y. Optical surface waves in periodic layered media. Appl. Phys. Lett. 1978, 32, 104–105. [Google Scholar] [CrossRef]
- Meade, R.D.; Brommer, K.D.; Rappe, A.M.; Joannopoulos, J.D. Electromagnetic bloch waves at the surface of a photonic crystal. Phys. Rev. B 1991, 44, 10961–10964. [Google Scholar] [CrossRef]
- Robertson, W.M.; May, M.S. Surface electromagnetic wave excitation on one-dimensional photonic band gap arrays. Appl. Phys. Lett. 1999, 74, 1800–1802. [Google Scholar] [CrossRef]
- Shinn, M.; Robertson, W.M. Surface plasmon-like sensor based on surface electromagnetic waves in a photonic band gap material. Sens. Actuators B Chem. 2005, 105, 360–364. [Google Scholar] [CrossRef]
- Descrovi, E.; Frascella, F.; Sciacca, B.; Geobaldo, F.; Dominici, L.; Michelotti, F. Coupling of surface waves in highly defined one-dimensional porous silicon photonic crystals for gas sensing applications. Appl. Phys. Lett. 2007, 91, 241109-1–241109-3. [Google Scholar] [CrossRef]
- Sfez, T.; Descrovi, E.; Dominici, L.; Nakagawa, W.; Michelotti, F.; Giorgis, F.; Herzig, H.P. Near-field analysis of surface electromagnetic waves in the bandgap region of a polymeric grating written on a one-dimensional photonic crystal. Appl. Phys. Lett. 2008, 93, 061108-1–061108-3. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Danz, N.; Descrovi, E.; Munzertb, P.; Schulz, U.; Sonntag, F.; Dominici, L.; Michelotti, F. Direct comparison of the performance of bloch surface wave and surface plasmon polariton sensors. Sens. Actuators B Chem. 2012, 174, 292–298. [Google Scholar] [CrossRef]
- Li, Y.; Yang, T.; Pang, Z.; Du, G.; Song, S.; Han, S. Phase-sensitive bloch surface wave sensor based on variable angle spectroscopic ellipsometry. Opt. Expr. 2014, 22, 21403–21410. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Villeneuve, P.R.; Joannopoulos, J.D.; Schubert, E.F. High extraction efficiency of spontaneous emission from slabs of photonic crystals. Phys. Rev. Lett. 1997, 18, 3294–3297. [Google Scholar] [CrossRef]
- Kanskar, M.; Paddon, P.; Pacradouni, V.; Morin, R.; Busch, A.; Young, J.F.; Johnson, S.R.; MacKenzie, J.; Tiedje, T. Observation of leaky slab modes in an air-bridged semiconductor waveguide with a two-dimensional photonic lattice. Appl. Phys. Lett. 1997, 70, 1438–1440. [Google Scholar]
- Villeneuve, P.R.; Fan, S.; Johnson, S.G.; Joannopoulos, J.D. Three-dimensional photon confinement in photonic crystals of low-dimensional periodicity. IEEE Proc. Optoelectron. 1998, 145, 384–390. [Google Scholar] [CrossRef]
- Johnson, S.G.; Fan, S.; Villeneuve, P.R.; Joannopoulos, J.D.; Kolodziejski, L.A. Guided modes in photonic crystal slabs. Phys. Rev. B 1999, 60, 5751–5758. [Google Scholar] [CrossRef]
- Painter, O.; Vuckovic, J.; Scherer, A. Defect modes of a two-dimensional photonic crystal in an optically thin dielectric slab. J. Opt. Soc. Am. B 1999, 16, 275–285. [Google Scholar] [CrossRef]
- Boroditsky, M.; Vrijen, R.; Krauss, T.F.; Coccioli, R.; Bhat, R.; Yablonovitch, E. Spontaneous emission extraction and purcell enhancement from thin-film 2-d photonic crystals. Lightwave Technol. 1999, 17, 2096–2112. [Google Scholar] [CrossRef]
- Astratov, V.N.; Culshaw, I.S.; Stevenson, R.M.; Whittaker, D.M.; Skolnick, M.S.; Krauss, T.F.; de la Rue, R.M. Resonant coupling of near-infrared radiation to photonic band structure waveguides. Lightwave Technol. 1999, 17, 2050–2057. [Google Scholar] [CrossRef]
- Baba, T.; Fukaya, N.; Yonekura, J. Observation of light propagation in photonic crystal optical waveguides with bends. Electron. Lett. 1999, 35, 654–655. [Google Scholar] [CrossRef]
- Paddon, P.; Young, J.F. Two-dimensional vector-coupled-mode theory for textured planar waveguides. Phys. Rev. B 2000, 61, 2090–2101. [Google Scholar] [CrossRef]
- Pacradouni, V.; Mandeville, W.J.; Cowan, A.R.; Paddon, P.; Young, J.F.; Johnson, S.R. Photonic band structure of dielectric membranes periodically textured in two dimensions. Phys. Rev. B 2000, 62, 4204–4207. [Google Scholar] [CrossRef]
- Kuchinsky, S.; Allan, D.C.; Borrelli, N.F.; Cotteverte, J.C. 3D localization in a channel waveguide in a photonic crystal with 2d periodicity. Opt. Commun. 2000, 175, 147–152. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chow, E.; Johnson, S.G.; Joannopoulos, J.D. Demonstration of highly efficient waveguiding in a photonic crystal slab at the 1.5-um wavelength. Opt. Lett. 2000, 25, 1297–1299. [Google Scholar] [CrossRef] [PubMed]
- Benisty, H.; Labilloy, D.; Weisbuch, C.; Smith, C.J.M.; Krauss, T.F.; Cassagne, D.; Beraud, A.; Jouanin, C. Radiation losses of waveguide-based two-dimensional photonic crystals: Positive role of the substrate. Appl. Phys. Lett. 2000, 76, 532–534. [Google Scholar] [CrossRef]
- Chutinan, A.; Noda, S. Waveguides and waveguide bends in two-dimensional photonic crystal slabs. Phys. Rev. B 2000, 62, 4488–4492. [Google Scholar] [CrossRef]
- Cowan, A.R.; Paddon, P.; Pacradouni, V.; Young, J.F. Resonant scattering and mode coupling in two-dimensional textured planar waveguides. J. Opt. Soc. Am. A 2001, 18, 1160–1170. [Google Scholar] [CrossRef]
- Vahala, K. Optical Microcavities (Advanced Series in Applied Physics), 1st ed.; World Scientifc Pubishing Company: Singapore, 2004. [Google Scholar]
- Block, I.D.; Mathias, P.C.; Ganesh, N.; Jones, I.D.; Dorvel, B.R.; Chaudhery, V.; Vodkin, L.; Bashir, R.; Cunningham, B.T. A detection instrument for enhanced fluorescence and label-free imaging on photonic crystal surfaces. Opt. Expr. 2009, 17, 13222–13235. [Google Scholar] [CrossRef]
- Schulz, S.C. Web based photonic crystal biosensors for drug discovery & diagnostics. Vac. Coat. 2008, 68. [Google Scholar]
- Krebs, F.C. Polymer solar cell modules prepared using roll-to-roll methods: Knife-over-edge coating, slot-die coating and screen printing. Sol. Energy Mater. Sol. Cells 2009, 93, 465–475. [Google Scholar] [CrossRef]
- Ge, C.; Lu, M.; Jian, X.; Tan, Y.F.; Cunningham, B.T. Large-area organic distributed feedback laser fabricated by nanoreplica molding and horizontal dipping. Opt. Expr. 2010, 18, 12980–12991. [Google Scholar] [CrossRef] [PubMed]
- Block, I.D.; Ganesh, N.; Lu, M.; Cunningham, B.T. A sensitivity model for predicting photonic crystal biosensor performance. IEEE Sens. J. 2008, 8, 274–280. [Google Scholar] [CrossRef]
- Block, I.D.; Mathias, P.C.; Jones, S.I.; Vodkin, L.O.; Cunningham, B.T. Optimizing the spatial resolution of photonic crystal label-free imaging. Appl. Opt. 2009, 48, 6567–6574. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.J.; Cunningham, B.T. Single-step fabrication and characterization of photonic crystal biosensors with polymer microfluidic channels. Lab Chip 2006, 6, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Mathias, P.C.; Ganesh, N.; Chan, L.L.; Cunningham, B.T. Combined enhanced fluorescence and label-free biomolecular detection with a photonic crystal surface. Appl. Opt. 2007, 46, 2351–2360. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.J.; Belobraydich, A.R.; Chan, L.L.; Mathias, P.C.; Cunningham, B.T. Comparison of label-free biosensing in microplate, microfluidic, and spot-based affinity capture assays. Anal. Biochem. 2010, 405, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, N.; Zhang, W.; Mathias, P.C.; Chow, E.; Soares, J.A.; Malyarchuk, V.; Smith, A.D.; Cunningham, B.T. Enhanced fluorescence emission from quantum dots on a photonic crystal surface. Nat. Nanotechnol. 2007, 2, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, N.; Block, I.D.; Mathias, P.C.; Zhang, W.; Chow, E.; Malyarchuk, V.; Cunningham, B.T. Leaky-mode assisted fluorescence extraction: Application to fluorescence enhancement biosensors. Opt. Expr. 2008, 16, 21626–21640. [Google Scholar] [CrossRef]
- Ganesh, N.; Mathias, P.C.; Zhang, W.; Cunningham, B.T. Distance dependence of fluorescence enhancement from photonic crystal surfaces. J. Appl. Phys. 2008, 103, 083104. [Google Scholar] [CrossRef]
- Pokhriyal, A.; Lu, M.; Huang, C.S.; Schulz, S.; Cunningham, B.T. Multicolor fluorescence enhancement from a photonics crystal surface. Appl. Phys. Lett. 2010, 97, 121108. [Google Scholar] [CrossRef] [PubMed]
- Pokhriyal, A.; Lu, M.; Chaudhery, V.; Huang, C.S.; Schulz, S.; Cunningham, B.T. Photonic crystal enhanced fluorescence using a quartz substrate to reduce limits of detection. Opt. Expr. 2010, 18, 24793–24808. [Google Scholar] [CrossRef] [PubMed]
- Mathias, P.C.; Ganesh, N.; Zhang, W.; Cunningham, B.T. Graded wavelength one-dimensional photonic crystal reveals spectral characteristics of enhanced fluorescence. J. Appl. Phys 2008, 103, 094320. [Google Scholar] [CrossRef]
- Mathias, P.C.; Wu, H.Y.; Cunningham, B.T. Employing two distinct photonic crystal resonances to improve fluorescence enhancement. Appl. Phys. Lett. 2009, 95, 201111. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Zhang, W.; Mathias, P.C.; Cunningham, B.T. Magnification of photonic crystal fluorescence enhancement via tm resonance excitation and te resonance extraction on a dielectric nanorod surface. Nanotechnology 2010, 21, 125203. [Google Scholar] [CrossRef] [PubMed]
- Chaudhery, V.; Lu, M.; Pokhriyal, A.; Schulz, S.C.; Cunningham, B.T. Angle-scanning photonic crystal enhanced fluorescence microscopy. IEEE Sens. J. 2012, 12, 1272–1279. [Google Scholar] [CrossRef]
- George, S.; Chaudhery, V.; Lu, M.; Takagi, M.; Amro, N.; Pokhriyal, A.; Tan, Y.F.; Ferreira, P.; Cunningham, B.T. Sensitive detection of protein and mirna cancer biomarkers using silicon-based photonic crystals and a resonance coupling laser scanning platform. Lab Chip 2013, 13, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Pokhriyal, A.; Lu, M.; Ge, C.; Cunningham, B.T. Coupled external cavity photonic crystal enhanced fluorescence. J. Biophotonics 2014, 7, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Long, K.D.; Yu, H.J.; Tan, Y.F.; Choi, J.S.; Harley, B.A.; Cunningham, B.T. Enhanced live cell imaging via photonic crystal enhanced fluorescence microscopy. Analyst 2014, 139, 5954–5963. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Mohseni, P.K.; Pan, L.; Li, X.; Somnath, S.; Felts, J.R.; Shannon, M.A.; King, W.P. Fabrication of arbitrarily-shaped silicon and silicon oxide nanostructures using tip-based nanofabrication. J. Vac. Sci. Technol. B 2013, 31, 06FJ01. [Google Scholar] [CrossRef]
- King, W.P.; Bhatia, B.; Felts, J.R.; Kim, H.J.; Kwon, B.; Lee, B.; Somnath, S.; Rosenberger, M. Heated atomic force microscope cantilevers and their applications. Annu. Rev. Heat Transf. 2013, 16, 287–326. [Google Scholar] [CrossRef]
- Tian, L.; Chen, E.; Gandra, N.; Abbas, A.; Singamaneni, S. Gold nanorods as plasmonic nanotransducers: Distance-dependent refractive index sensitivity. Langmuir 2012, 28, 17435–17442. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Morrissey, J.J.; Kattumenu, R.; Gandra, N.; Kharasch, E.D.; Singamaneni, S. Bioplasmonic paper as a platform for detection of kidney cancer biomarkers. Anal. Chem. 2012, 84, 9928–9934. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, Y.; Cunningham, B.T. Label-Free Biosensor Imaging on Photonic Crystal Surfaces. Sensors 2015, 15, 21613-21635. https://doi.org/10.3390/s150921613
Zhuo Y, Cunningham BT. Label-Free Biosensor Imaging on Photonic Crystal Surfaces. Sensors. 2015; 15(9):21613-21635. https://doi.org/10.3390/s150921613
Chicago/Turabian StyleZhuo, Yue, and Brian T. Cunningham. 2015. "Label-Free Biosensor Imaging on Photonic Crystal Surfaces" Sensors 15, no. 9: 21613-21635. https://doi.org/10.3390/s150921613




