Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Image Acquisition and Analysis
2.3. Yield and Quality Measurement
2.4. Nitrogen Concentration Determination
2.5. Nitrogen Nutrition Index (NNI) Calculation
2.6. Model Development
2.6.1. Feature Selection
2.6.2. Models for Predicting Plant N Nutrition
2.6.3. Model Validation
2.7. Model Evaluation in Different Scenarios
3. Results
3.1. Yield and Quality
3.2. Accumulation of Biomass and N Concentration
3.3. Nitrogen Nutrition Index
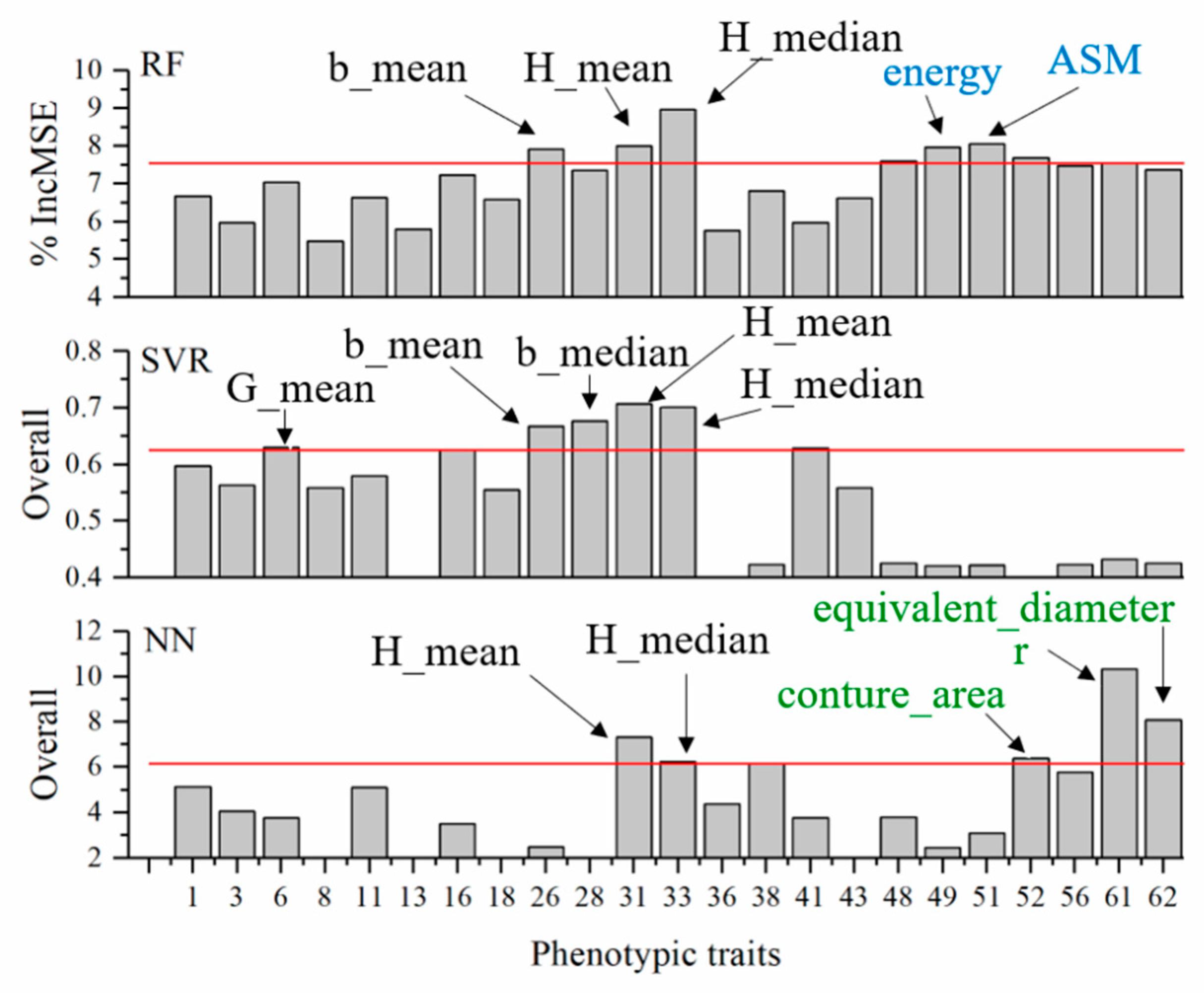
3.4. Feature Selection
3.5. Model Development
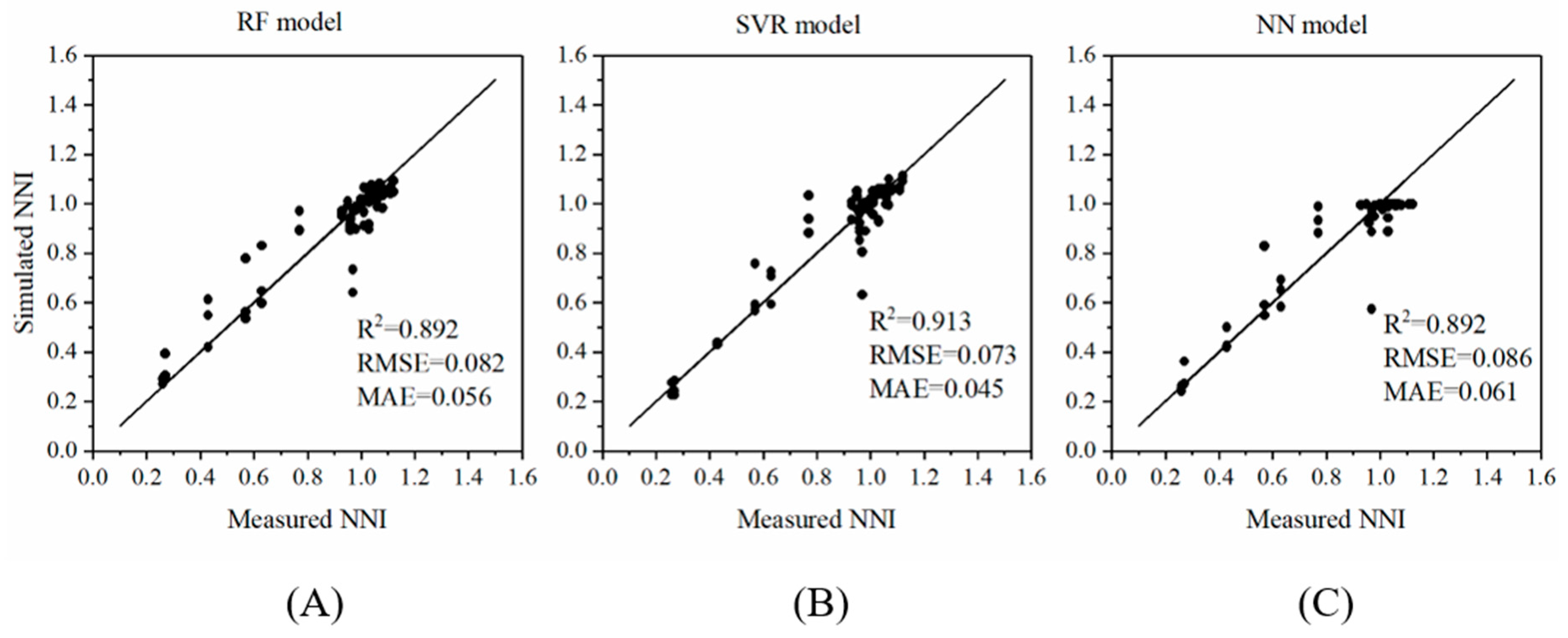
3.6. Model Validation
3.7. Model Evaluation
3.7.1. Model Evaluation under Different Plant N Nutrition Status
3.7.2. Model Evaluation under Different Growth Stages of Pakchoi
3.8. Relative Importance of Phenotypic Traits in Predicting Plant NNI
4. Discussion
4.1. Strategies for Integral Control of Water and Fertilization for Crops to Improve N Absorption and Utilization
4.2. Accuracy Comparison of Full Model with the Color Model in Predicting Plant Nitrogen
4.3. Random Forest (RF) Model based on Phenotyping Is the Most Robust in Predicting NNI
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yan, X.; Ti, C.; Vitousek, P.; Chen, D.; Leip, A.; Cai, Z.; Zhu, Z. Fertilizer nitrogen recovery efficiencies in crop production systems of China with and without consideration of the residual effect of nitrogen. Environ. Res. Lett. 2014, 9, 095002. [Google Scholar] [CrossRef] [Green Version]
- Nachmansohn, J. Minimized nutrient leaching through fertilizer management. Master’s Thesis, Swedish University of Agricultural Sciences, Uppsala, Swedish, 2016. [Google Scholar]
- Chen, Q.; Zhang, X.; Zhang, H.; Christie, P.; Li, X.; Horlacher, D.; Liebig, H.P. Evaluation of current fertilizer practice and soil fertility in vegetable production in the Beijing region. Nutr. Cycl. Agroecosyst. 2004, 69, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Gao, N.; Liu, Y.; Wu, H.; Zhang, P.; Yua, N.; Zhang, Y.; Zou, H.; Fan, Q.; Zhang, Y. Interactive effects of irrigation and nitrogen fertilizer on yield, nitrogen uptake, and recovery of two successive Chinese cabbage crops as assessed using 15 N isotope. Sci. Hortic. 2017, 215, 117–125. [Google Scholar] [CrossRef]
- Greenwood, D.J.; Lemaire, G.; Gosse, G.; Cruz, P.C.S.; Neeteson, J.J. Decline in percentage N of C3 and C4 crops with increasing plant mass. Ann. Bot. 1990, 66, 425–436. [Google Scholar] [CrossRef]
- He, Z.; Qiu, X.; Ata-Ul-karim, S.T.; Li, Y.; Liu, X.; Cao, Q.; Zhu, Y.; Cao, W.; Tang, L. Development of a critical nitrogen dilution curve of double cropping rice in south China. Front. Plant Sci. 2017, 8, 638. [Google Scholar] [CrossRef] [PubMed]
- Sedlář, O.; Balík, J.; Černý, J.; Kulhánek, M.; Vašák, F. Relation between nitrogen nutrition index and production of spring malting barley. Int. J. Plant Prod. 2017, 379–388. [Google Scholar] [CrossRef]
- Yin, M.; Li, Y.; Xu, L.; Shen, S.; Fang, H. Nutrition diagnosis for ditrogen in winter wheat based on critical nitrogen dilution curves. Crop Sci. 2017, 58, 416–425. [Google Scholar] [CrossRef]
- Zhao, B.; Ata-Ul-Karim, S.T.; Liu, Z.; Ning, D.; Xiao, J.; Liu, Z.; Nan, J.; Duan, A. Development of a critical nitrogen dilution curve based on leaf dry matter for summer maize. Field Crop. Res. 2017, 208, 60–68. [Google Scholar] [CrossRef]
- Lemaire, G.; Gastal, F. N uptake and distribution in plant Canopies; Springer: Berlin/Heidelberg, Germany, 1997; pp. 3–43. [Google Scholar]
- Fox, R.H.; Walthall, C.L. Crop Monitoring technologies to assess nitrogen status. Nitrogen Agric. Syst. 2008, nitrogeninagric, 647–674. [Google Scholar]
- Ata-Ul-karim, S.T.; Cao, Q.; Qiang, C.; Yan, Z.; Liang, T.; Asif, R.M.I.; Weixing, C. Non-destructive assessment of plant nitrogen parameters using leaf chlorophyll measurements in rice. Front. Plant Sci. 2016, 7, 1829. [Google Scholar] [CrossRef]
- Ravier, C.; Quemada, M.; Jeuffroy, M.H. Use of a chlorophyll meter to assess nitrogen nutrition index during the growth cycle in winter wheat. Field Crop. Res. 2017, 214, 73–82. [Google Scholar] [CrossRef]
- Yang, H.; Yang, J.P.; Lv, Y.M.; He, J.J. Replacing the nitrogen nutrition index by SPAD values and analysis of effect factors for estimating rice nitrogen status. Agron. J. 2018, 110, 545–554. [Google Scholar] [CrossRef]
- Nigon, T.J.; Mulla, D.J.; Rosen, C.J.; Cohen, Y.; Alchanatis, V.; Knight, J.; Rud, R. Hyperspectral aerial imagery for detecting nitrogen stress in two potato cultivars. Comput. Electron. Agr. 2015, 112, 36–46. [Google Scholar] [CrossRef]
- Feng, M.; Zhao, J.; Yang, W.; Wang, C.; Zhang, M.; Xiao, L.; Ding, G. Evaluating winter wheat (Triticum aestivum L.) nitrogen status with canopy spectrum reflectance and multiple statistical analysis. Spectrosc. Lett. 2016, 49, 507–513. [Google Scholar] [CrossRef]
- Chen, L.; Lin, L.; Cai, G.; Sun, Y.; Huang, T.; Wang, K.; Deng, J. Identification of nitrogen, phosphorus, and potassium deficiencies in rice based on static scanning technology and hierarchical identification method. PLoS One 2014, 9, e113200. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Zhang, Y.; Tremblay, N.; Zhu, J. A first comparison of Multiplex® for the assessment of corn nitrogen status. J. Food. Agric. Environ. 2012, 10, 1008–1016. [Google Scholar]
- Agati, G.; Foschi, L.; Guglielminetti, L.; Volterrani, M.; Grossi, N.; Cerovic, Z.G. Fluorescence-based versus reflectance proximal sensing of nitrogen content in Paspalum vaginatum and Zoysia matrella turfgrasses. Eur. J. Agron. 2013, 45, 39–51. [Google Scholar] [CrossRef]
- Agati, G.; Foschi, L.; Grossi, N.; Volterrani, M. In field non-invasive sensing of the nitrogen status in hybrid bermudagrass (Cynodon dactylon × C. transvaalensis Burtt Davy) by a fluorescence-based method. Eur. J. Agron. 2015, 63, 89–96. [Google Scholar] [CrossRef]
- Padilla, F.M.; Peña-Fleitas, M.T.; Gallardo, M.; Thompson, R.B. Evaluation of optical sensor measurements of canopy reflectance and of leaf flavonols and chlorophyll contents to assess crop nitrogen status of muskmelon. Eur. J. Agron. 2014, 58, 39–52. [Google Scholar] [CrossRef]
- Agati, G.; Tuccio, L.; Kusznierewicz, B.; Chmiel, T.; Bartoszek, A.; Kowalski, A.; Grzegorzewska, A.; Kosson, R.; Kaniszewski, S. Nondestructive optical sensing of flavonols and chlorophyll in white head cabbage (Brassica oleracea L. var. capitata subvar. alba) grown under different nitrogen regimens. J. Agric. Food Chem. 2015, 64, 85–94. [Google Scholar] [CrossRef]
- Poiré, R.; Chochois, V.; Sirault, X.R.R.; Vogel, J.P.; Furbank, R.T. Digital imaging approaches for phenotyping whole plant nitrogen and phosphorus response in Brachypodium distachyon. J. Integr. Plant Biol. 2014, 56, 781–796. [Google Scholar] [CrossRef]
- Baresel, J.P.; Rischbeck, P.; Hu, Y.; Kipp, S.; Gero, B.; Mistele, B.; Schmidhalter, U. Use of a digital camera as alternative method for non-destructive detection of the leaf chlorophyll content and the nitrogen nutrition status in wheat. Comput. Electron. Agr. 2017, 140, 25–33. [Google Scholar] [CrossRef]
- Sun, Y.; Tong, C.; He, S.; Wang, K.; Chen, L. Identification of nitrogen, phosphorus, and potassium deficiencies based on temporal dynamics of leaf morphology and color. Sustainability. 2018, 10, 762–775. [Google Scholar] [CrossRef]
- Guerrero, J.M.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for crop/weeds identification in maize fields. Expert Syst. Appl. 2012, 39, 39. [Google Scholar] [CrossRef]
- Sonnentag, O.; Hufkens, K.; Teshera-Sternea, C.; Young, A.M.; Friedl, M.; Braswell, B.H.; Milliman, T.; O’Keefe, J.; Richardson, A.D. Digital repeat photography for phenological research in forest ecosystems. Agr. Forest. Meteorol. 2012, 152, 159–177. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, G.; Wang, J. Estimating nitrogen status of rice using the image segmentation of G-R thresholding method. Field Crop. Res. 2013, 149, 33–39. [Google Scholar] [CrossRef]
- Li, J.; Shi, Y.; Veeranampalayam-Sivakumar, A.N.; Schachtman, D.P. Elucidating sorghum biomass, nitrogen and chlorophyll contents with spectral and morphological traits derived from unmanned aircraft system. Front. Plant Sci. 2018, 9, 1406. [Google Scholar] [CrossRef]
- Duan, L.F.; Han, J.W.; Guo, Z.L.; Tu, H.F.; Yang, P.; Zhang, D.; Fan, Y.; Chen, G.X.; Xiong, L.Z.; Dai, M.Q.; Williams, K.; Corke, F.; Doonan, J.H.; Yang, W.N. Novel digital features discriminate between drought resistant and drought sensitive rice under controlled and field conditions. Front. Plant Sci. 2018, 9, 492. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.J.; Burgos-Artizzu, X.P.; Ribeiro, A. Automatic segmentation of relevant textures in agricultural images. Comput. Electron. Agr. 2011, 75, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Torres-Sánchez, J.; Pena, J.M.; De Castro, A.I.; López-Granados, F. Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Comput. Electron. Agr. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Marcial-Pablo, M.D.J.; Gonzalez-Sanchez, A.; Jimenez-Jimenez, S.I.; Ontiveros-Capurata, R.E.; Ojeda-Bustamante, W. Estimation of vegetation fraction using RGB and multispectral images from UAV. Int. J. Remote Sens. 2019, 40, 420–438. [Google Scholar] [CrossRef]
- Gehan, M.A.; Fahlgren, N.; Abbasi, A.; Berry, J.C.; Sax, T. PlantCV v2: Image analysis software for high-throughput plant phenotyping. Peer J. 2017, 5, e4088. [Google Scholar] [CrossRef]
- Perez-Sanz, F.; Navarro, P.J.; Egea-Cortines, M. Plant phenomics: an overview of image acquisition technologies and image data analysis algorithms. GigaScience 2017, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Sulistyo, S.B.; Woo, W.L.; Dlay, S.; Gao, B. Building a globally optimized computational intelligent image processing algorithm for on-site nitrogen status analysis in plants. IEEE Intel. Syst. 2018. [Google Scholar] [CrossRef]
- Sulistyo, S.B.; Woo, W.L.; Dlay, S.S. Regularized neural networks fusion and genetic algorithm based on-field nitrogen status estimation of wheat plants. IEEE T. Ind. Inform. 2017, 13, 103–114. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef]
- Alvaro, F.; Sook, Y.; Sang, K.; Dong, P. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Informa. Process. Agr. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agr. 2018, S0, 168169917312619. [Google Scholar] [CrossRef]
- Guo, D.D.; Juan, J.X.; Chang, L.Y.; Zhang, J.J.; Huang, D.F. Discrimination of plant root zone water status in greenhouse production based on phenotyping and machine learning techniques. Sci. Rep. 2017, 7, 8303. [Google Scholar] [CrossRef]
- Gonzaacute, J.L.; Espino-Gudiño, M.C.; Gudiño-Bazaldúa, J.; Rojas-Rentería, J.L.; Rodríguez-Hernández, V.; Castaño, V.M. Color image segmentation using perceptual spaces through applets for determining and preventing diseases in chili peppers. African J. Biotechnol. 2013, 679–688. [Google Scholar] [CrossRef]
- Calderón, R.; Navascortés, J.A.; Pablo, Z.T. Early detection and quantification of verticillium wilt in olive using hyperspectral and thermal imagery over large areas. Remote Sens. 2015, 7, 5584–5610. [Google Scholar] [CrossRef]
- Yu, X.; Liu, T.; Zheng, X.; Yang, Z.; Wang, J. Prediction of regulatory interactions in Arabidopsis using gene-expression data and support vector machines. Plant Physiol. Bioch. 2011, 49, 280–283. [Google Scholar] [CrossRef]
- Du, H.; Wang, J.; Hu, Z.; Yao, X.; Zhang, X. Prediction of fungicidal activities of rice blast disease based on least-squares support vector machines and project pursuit regression. J. Agr. Food Chem. 2008, 56, 10785–10792. [Google Scholar] [CrossRef]
- Cavallo, D.P.; Cefola, M.; Pace, B.; Logrieco, A.F.; Attolico, G. Contactless and non-destructive chlorophyll content prediction by random forest regression: A case study on fresh-cut rocket leaves. Comput. Electron. Agr. 2017, 140, 303–310. [Google Scholar] [CrossRef]
- Unseok, L.; Sungyul, C.; Anantrio, P.G.; Hyoungseok, K.; Hwan, K.D.; Hector, C. An automated, high-throughput plant phenotyping system using machine learning-based plant segmentation and image analysis. PLoS One 2018, 13, e0196615. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Xiong, X.; Chang, L.Y.; Muhammad, K.; Zhang, J.J.; Huang, D.F. Alleviation of drought stress by nitrogen application in Brassica campestris ssp. Chinensis L. Agronomy 2018, 8, 66. [Google Scholar] [CrossRef]
- Justes, E.; Mary, B.; Meynard, J.M.; Machet, J.M.; Thelier-Huche, L. Determination of a Critical Nitrogen Dilution Curve for Winter Wheat Crops. Ann. Bot. 1994, 74, 397–407. [Google Scholar] [CrossRef]
- Ziadi, N.; Brassard, M.; Bélanger, G.; Cambouris, A.N.; Tremblay, N.; Nolin, M.C.; Claessens, A.; Parent, L.E. Critical nitrogen curve and nitrogen nutrition index for corn in eastern Canada. Agrono. J. 2008, 100, 271–276. [Google Scholar] [CrossRef]
- Deming, S.N.; Michotte, Y.; Massart, D.L.; Kaufman, L.; Vandeginste, B.G.M. Chemometrics; Elsevier: Amsterdam, The Netherlands, 1988. [Google Scholar]
- Min, J.; Zhao, X.; Shi, W.M.; Xing, G.X.; Zhu, Z.L. Nitrogen balance and loss in a greenhouse vegetable system in southeastern China. Pedosphere 2011, 21, 464–472. [Google Scholar] [CrossRef]
- Ju, M.; Zhang, H.; Shi, W. Optimizing nitrogen input to reduce nitrate leaching loss in greenhouse vegetable production. Agr. Water Manage. 2012, 111, 53–59. [Google Scholar] [CrossRef]
- Rorie, R.L.; Purcell, L.C.; Morteza, M.; Douglas, K.E.; Andy, K.; Matthew, M.C.; David, L.E. Association of “greenness” in corn with yield and leaf nitrogen concentration. Agron. J. 2011, 103, 529–535. [Google Scholar] [CrossRef]
- Tei, F.; Nicola, S.; Benincasa, P. Advances in Research on Fertilization Management of Vegetable Crops; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Cartelat, A.; Cerovic, Z.G.; Goulas, Y.; Meyer, S.; Lelarge, C.; Prioul, J.L.; Barbottin, A.; Jeuffroy, M.H.; Gate, P.; Agati, G.; Moya, I. Optically assessed contents of leaf polyphenolics and chlorophyll as indicators of nitrogen deficiency in wheat (Triticum aestivum L.). Field Crop. Res. 2005, 91, 35–49. [Google Scholar] [CrossRef]
- Padilla, F.M.; Peña-Fleitas, M.T.; Morteza, M.; Douglas, K.E.; Andy, K.; Matthew, M.C.; David, L.E. Proximal optical sensing of cucumber crop N status using chlorophyll fluorescence indices. Eur. J. Agron. 2016, 73, 83–97. [Google Scholar] [CrossRef]

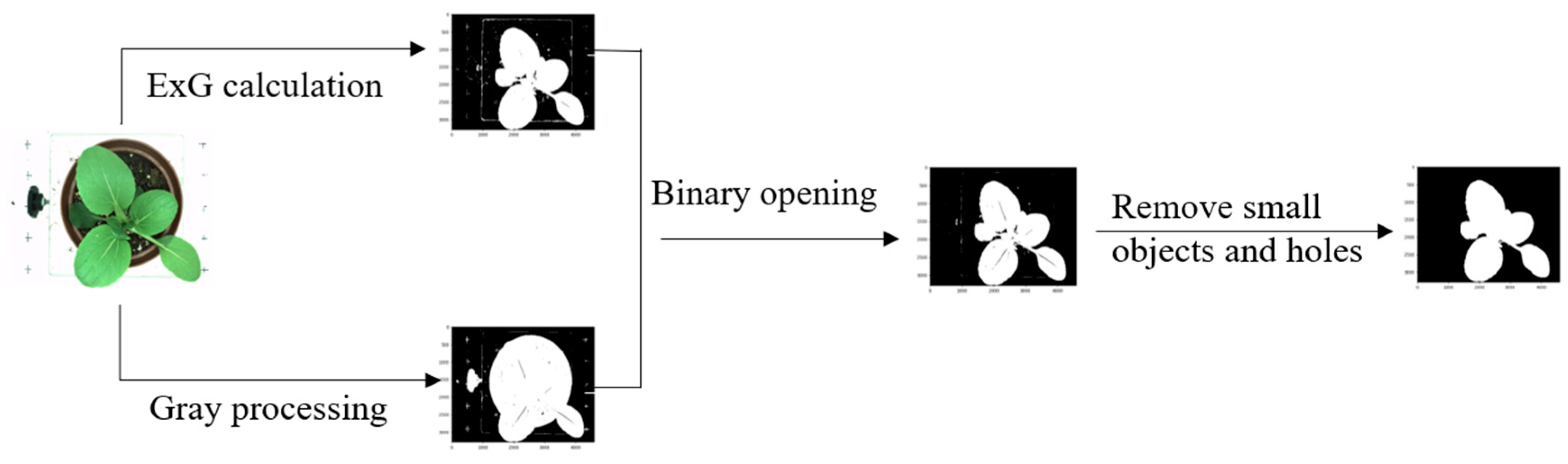
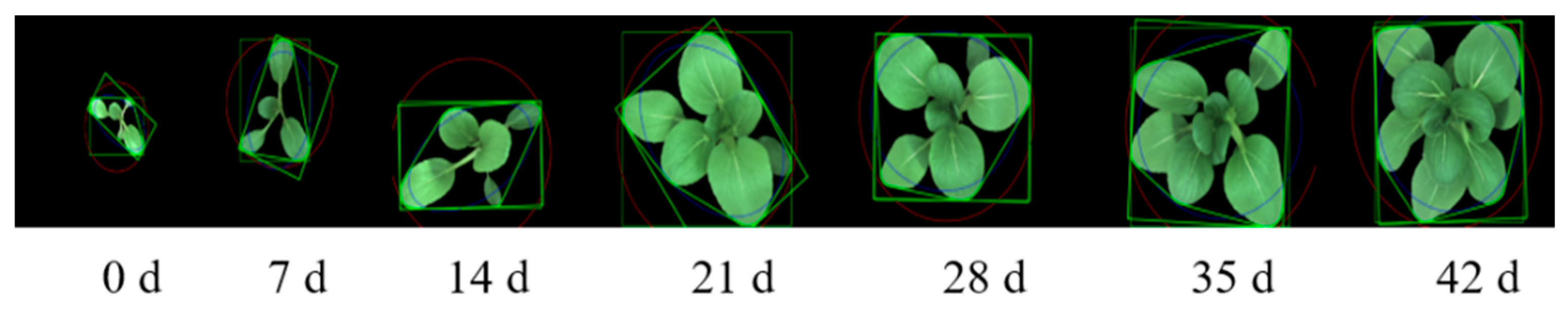
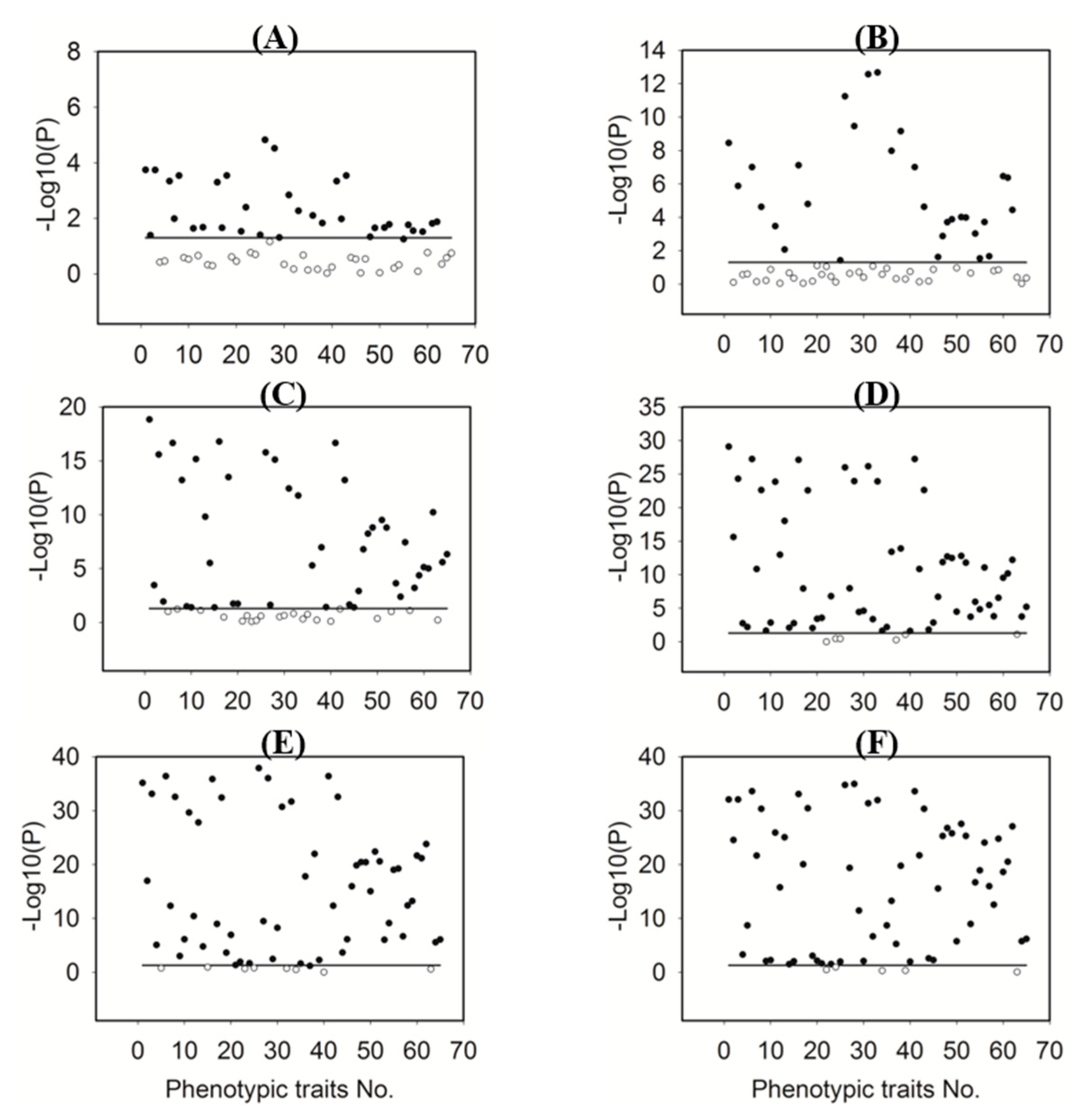

| Category | Trait Name | Description | No. |
|---|---|---|---|
| Color | *R_mean, R_std, *R_median, R_ range, R_coefficient of variation | Statistics in the red range of the RGB color space | 1–5 |
| *G_mean, G_std, *G_median, G_ range, G_coefficient of variation | Statistics in the green range of the RGB color space | 6–10 | |
| *B_mean, B_std, *B_median, B_ range, B_coefficient of variation | Statistics in the blue range of the RGB color space | 11–15 | |
| *L_mean, L_std, *L_median, L_ range, L_coefficient of variation | Statistics of L channel in LAB color space | 16–20 | |
| a_mean, a_std, a_median, a_ range, a_coefficient of variation | Statistics of A channel in LAB color space | 21–25 | |
| *b_mean, b_std, *b_median, b_ range, b_coefficient of variation | Statistics of B channel in LAB color space | 26–30 | |
| *H_mean, H_std, *H_median, H_ range, H_coefficient of variation | Statistics of H in HSV color space | 31–35 | |
| *S_mean, S_std, *S_median, S_ range, S_coefficient of variation | Statistics of S in HSV color space | 36–40 | |
| *V_mean, V_std, *V_median, V_ range, V_coefficient of variation | Statistics of V in HSV color space | 41–45 | |
| Texture | contrast | Definition and grooving depth of texture | 46 |
| dissimilarity | The difference of grey scale | 47 | |
| *homogeneity | The local changes of image texture | 48 | |
| *energy | The degree of thickness and uniformity for texture | 49 | |
| correlation | The correlation of the local grey scale | 50 | |
| *ASM | Angular second moment | 51 | |
| Morphology | *contour_area | Area of plant contour | 52 |
| perimeter | The length of plant contour | 53 | |
| w | The width of the bounding box | 54 | |
| h | The height of the bounding box | 55 | |
| *hull_area | Convex hull area (mm2) | 56 | |
| _w | The width of the minimum circumscribed rectangle | 57 | |
| _h | The height of the minimum circumscribed rectangle | 58 | |
| MA | The macro axis of the ellipse | 59 | |
| ma | The minor axis of the ellipse | 60 | |
| *r | The radius of the minimum circumscribed circle | 61 | |
| *equivalent_diameter | The diameter of a circle equal to the contour area | 62 | |
| aspect_ration | The width-height ratio of the bounding box | 63 | |
| extent | The area ratio between contour and bounding box | 64 | |
| solidity | The area ratio between contour and convex hull | 65 |
| Treatments | Yield (g·plant−1) | Chlorophyll (μg·cm−2) | Flavonol Index | Nitrate (mg·kg−1 FW) | Soluble Protein (mg·g−1 FW) |
|---|---|---|---|---|---|
| CK | 6.04 ± 0.45d | 28.70 ± 1.67b | 1.37 ± 0.26a | 200.92 ± 3.51c | 5.21 ± 0.58c |
| T1 | 27.65 ± 0.28a | 32.98 ± 4.98a | 0.74 ± 0.11b | 271.47 ± 23.35b | 35.10 ± 1.97a |
| T2 | 23.88 ± 0.04b | 32.17 ± 2.78a | 0.80 ± 0.11b | 298.31 ± 19.16b | 31.63 ± 1.91b |
| T3 | 21.37 ± 0.34c | 33.08 ± 2.75a | 0.78 ± 0.11b | 364.25 ± 25.34a | 30.47 ± 2.61b |
| Index | Treatment | Days after Transplant (d) | |||||
|---|---|---|---|---|---|---|---|
| 7 | 14 | 21 | 28 | 35 | 42 | ||
| Biomass | CK | 0.065 ± 0.007b | 0.161 ± 0.006b | 0.186 ± 0.008c | 0.338 ± 0.012c | 0.479 ± 0.037b | 0.617 ± 0.037c |
| T1 | 0.067 ± 0.004b | 0.174 ± 0.019b | 0.293 ± 0.010b | 0.580 ± 0.023b | 0.973 ± 0.019a | 1.363 ± 0.032a | |
| T2 | 0.080 ± 0.004a | 0.189 ± 0.023b | 0.306 ± 0.014b | 0.574 ± 0.018b | 0.994 ± 0.051a | 1.280 ± 0.025b | |
| T3 | 0.079 ± 0.010a | 0.215 ± 0.011a | 0.440 ± 0.025a | 0.620 ± 0.014a | 0.970 ± 0.017a | 1.282 ± 0.027b | |
| Nitrogen concentration | CK | 43.30 ± 2.31b | 39.70 ± 2.76b | 39.07 ± 1.59b | 30.80 ± 1.57d | 20.50 ± 1.08c | 20.50 ± 0.79c |
| T1 | 57.50 ± 0.26a | 61.27 ± 1.01a | 69.63 ± 1.90a | 67.30 ± 0.17c | 72.70 ± 2.66b | 83.77 ± 2.57b | |
| T2 | 57.63 ± 0.85a | 62.20 ± 1.23a | 74.87 ± 2.25a | 71.97 ± 0.99b | 81.87 ± 1.65a | 88.07 ± 2.67ab | |
| T3 | 60.57 ± 1.27a | 64.00 ± 0.85a | 73.30 ± 4.98a | 76.63 ± 0.50a | 79.47 ± 0.74a | 88.80 ± 2.11a | |
| Treatment | Days after Transplant (d) | |||||
|---|---|---|---|---|---|---|
| 7 | 14 | 21 | 28 | 35 | 42 | |
| CK | 0.77 | 0.63 | 0.57 | 0.43 | 0.27 | 0.26 |
| T1 | 1.03 | 0.97 | 1.01 | 0.93 | 0.95 | 1.05 |
| T2 | 1.03 | 0.98 | 1.08 | 1.00 | 1.07 | 1.11 |
| T3 | 1.08 | 1.01 | 1.06 | 1.06 | 1.04 | 1.12 |
| Different Scenarios | Range of Measured NNI | Model | Model Evaluation Results | |||
|---|---|---|---|---|---|---|
| Range of Simulated NNI | R2 | Relative Error (%) | Accuracy | |||
| Excessive | 1.01~1.12 | RF | 0.895~1.101 | 0.470 | 2.57 | 0.787 |
| SVR | 0.772~1.135 | 0.206 | 3.56 | 0.586 | ||
| NN | 0.822~1.000 | 0.016 | 1.76 | 0.085 | ||
| Low | 0.26~0.93 | RF | 0.262~1.081 | 0.945 | 8.35 | 0.948 |
| SVR | 0.173~1.087 | 0.921 | 10.51 | 0.984 | ||
| NN | 0.237~1.000 | 0.918 | 10.35 | 0.952 | ||
| Seedling period | 0.63~1.08 | RF | 0.495~1.068 | 0.795 | 6.56 | 0.823 |
| SVR | 0.348~1.084 | 0.703 | 8.79 | 0.856 | ||
| NN | 0.416~1.000 | 0.674 | 8.35 | 0.766 | ||
| Harvest period | 0.26~1.12 | RF | 0.262~1.101 | 0.985 | 4.49 | 0.943 |
| SVR | 0.173~1.135 | 0.981 | 5.23 | 0.974 | ||
| NN | 0.237~1.000 | 0.969 | 6.06 | 0.869 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, X.; Zhang, J.; Guo, D.; Chang, L.; Huang, D. Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L. Sensors 2019, 19, 2448. https://doi.org/10.3390/s19112448
Xiong X, Zhang J, Guo D, Chang L, Huang D. Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L. Sensors. 2019; 19(11):2448. https://doi.org/10.3390/s19112448
Chicago/Turabian StyleXiong, Xin, Jingjin Zhang, Doudou Guo, Liying Chang, and Danfeng Huang. 2019. "Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L." Sensors 19, no. 11: 2448. https://doi.org/10.3390/s19112448
APA StyleXiong, X., Zhang, J., Guo, D., Chang, L., & Huang, D. (2019). Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L. Sensors, 19(11), 2448. https://doi.org/10.3390/s19112448





