Application of High-Throughput Screening Raman Spectroscopy (HTS-RS) for Label-Free Identification and Molecular Characterization of Pollen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
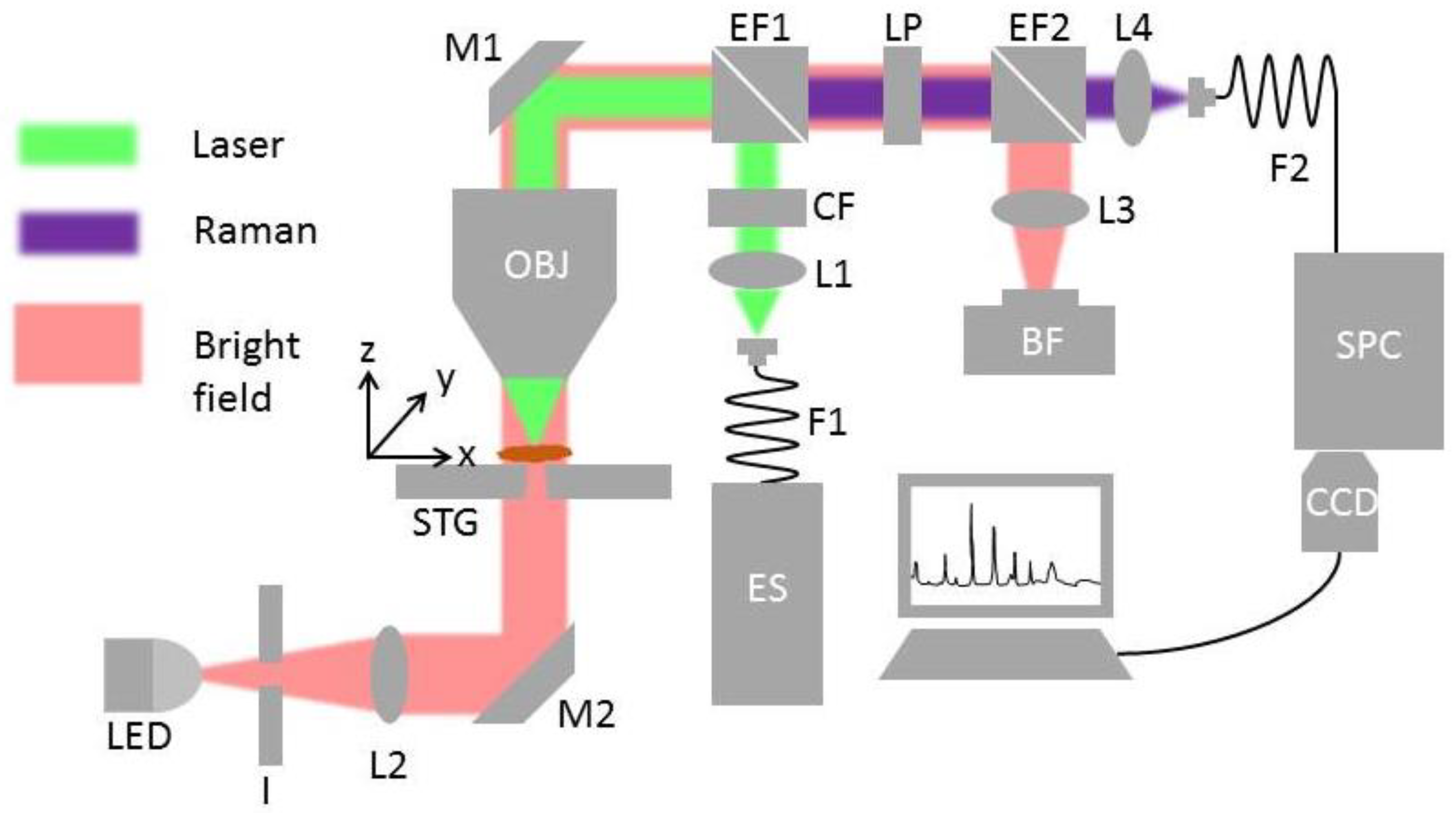
2.2. HTS-RS Platform
2.3. Raman Measurement of the Pollen Sample
2.4. Pollen Localization from Bright Field Image
2.5. Data Analysis of Raman Spectra
3. Results
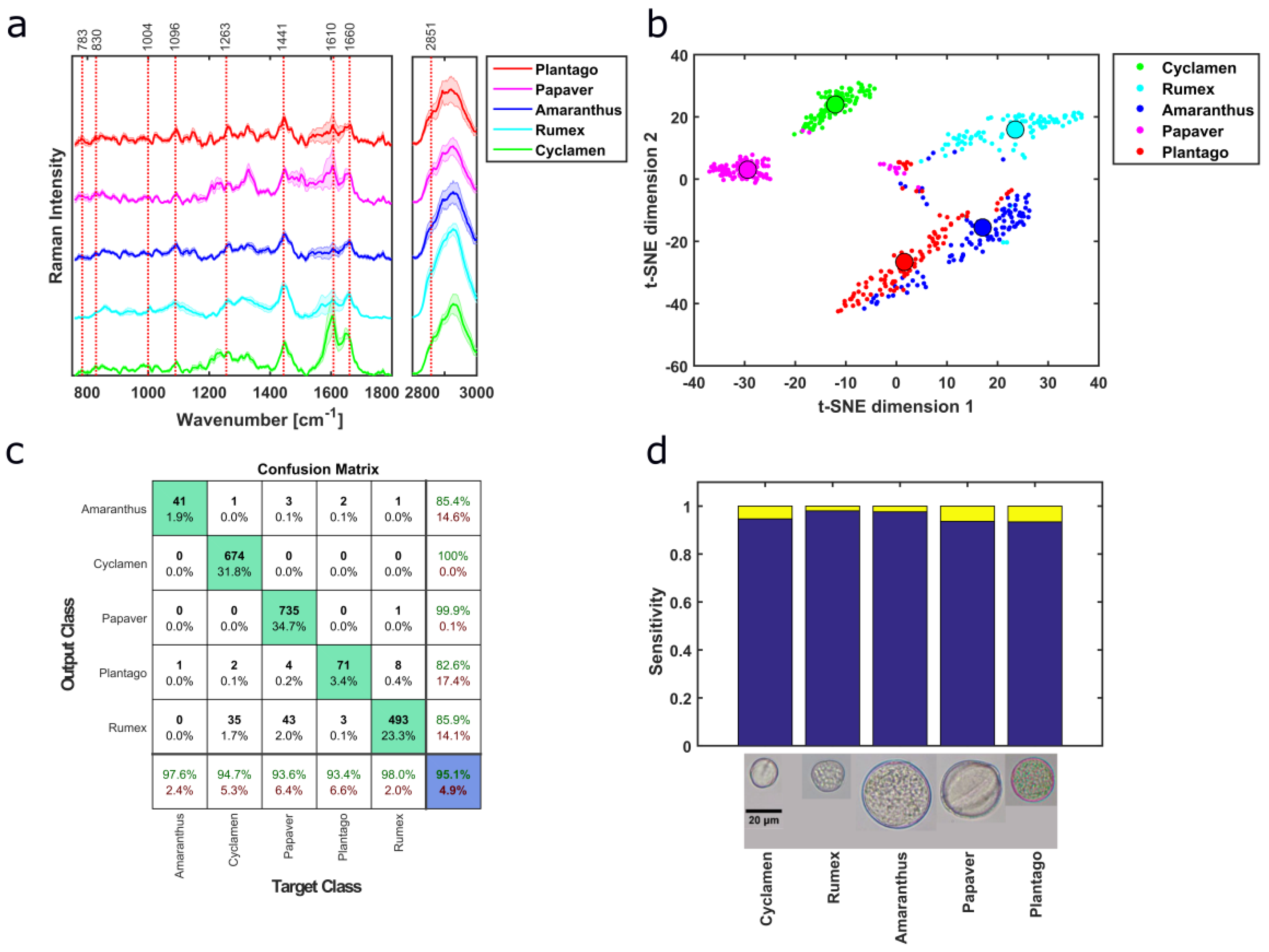
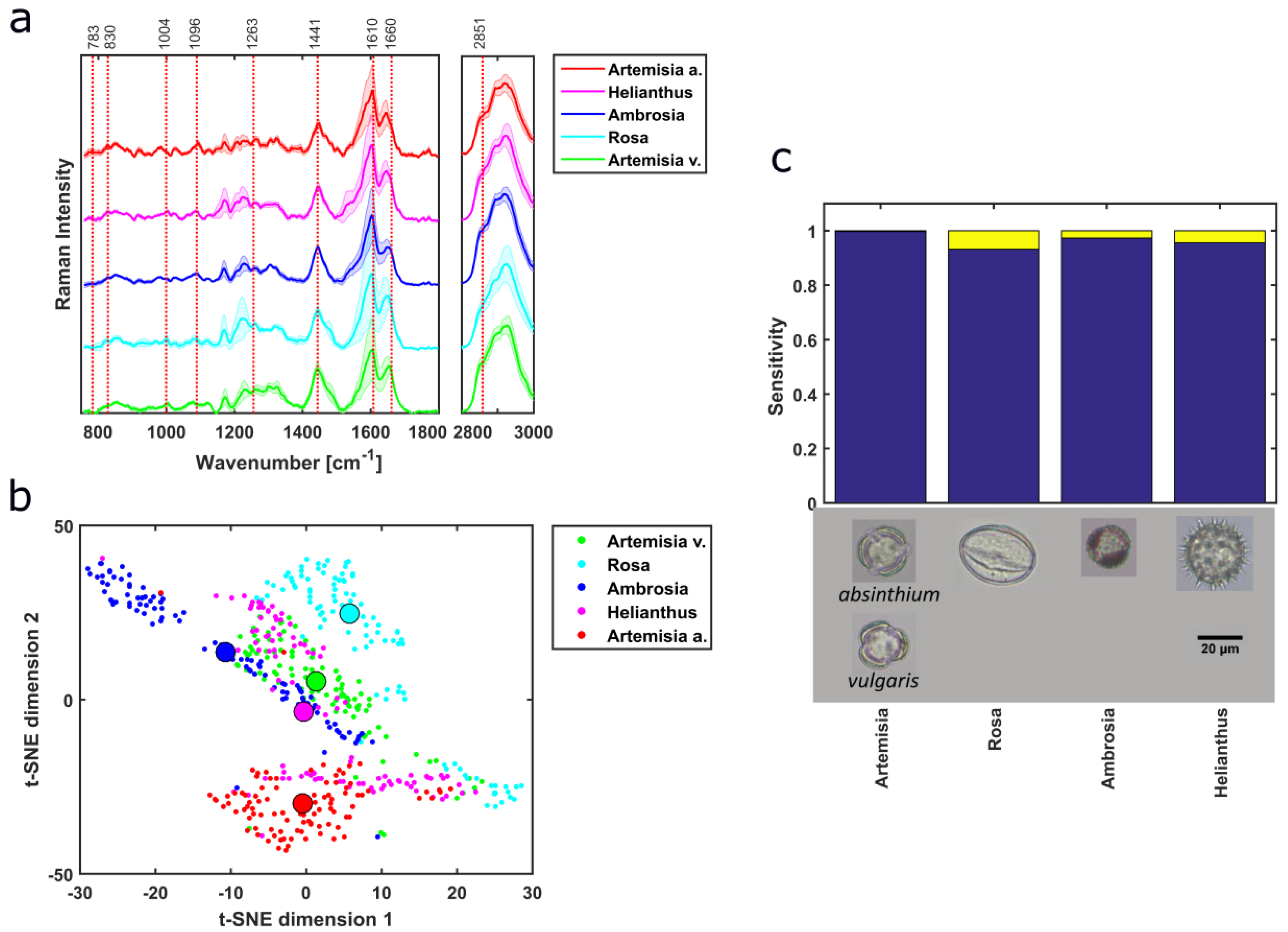
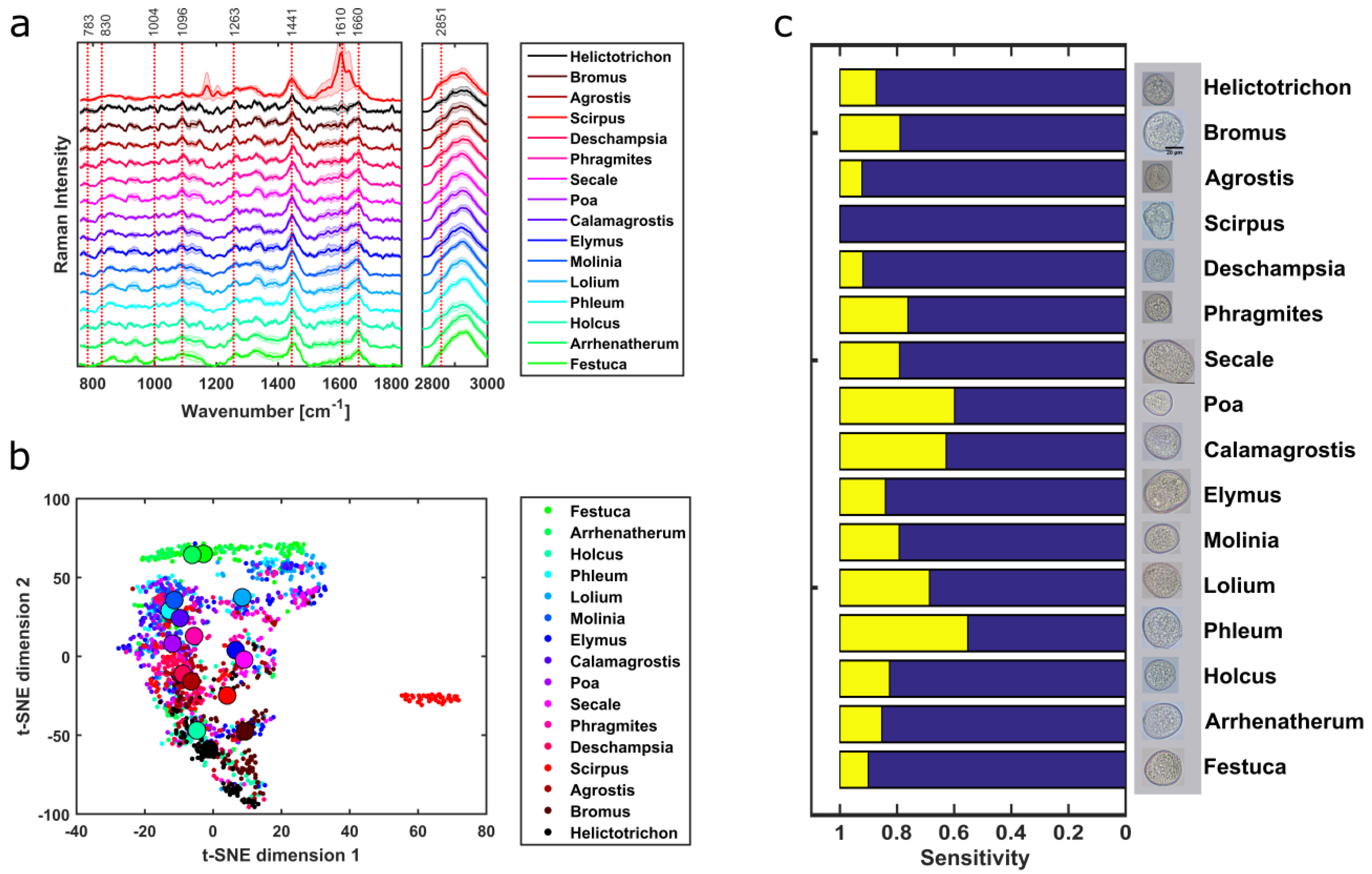
3.1. Pollen Type Identification Using Raman Spectra
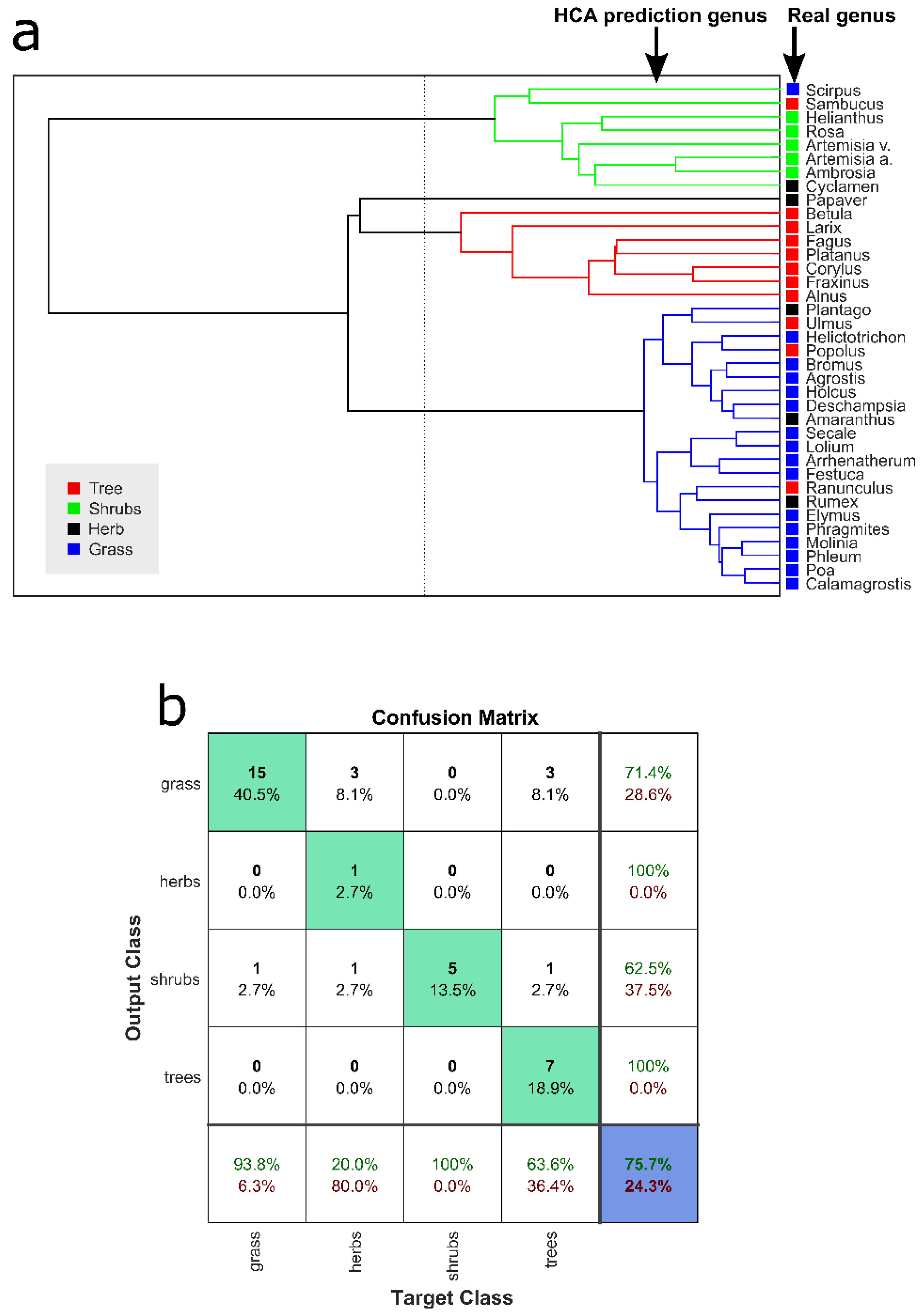
3.2. Taxonomic Discrimination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zimmermann, B.; Kohler, A. Infrared Spectroscopy of Pollen Identifies Plant Species and Genus as Well as Environmental Conditions. PLoS ONE 2014, 9, e95417. [Google Scholar] [CrossRef] [PubMed]
- Bağcıoğlu, M.; Zimmermann, B.; Kohler, A. A Multiscale Vibrational Spectroscopic Approach for Identification and Biochemical Characterization of Pollen. PLoS ONE 2015, 10, e0137899. [Google Scholar] [CrossRef] [PubMed]
- Gottardini, E.; Rossi, S.; Cristofolini, F.; Benedetti, L. Use of Fourier transform infrared (FT-IR) spectroscopy as a tool for pollen identification. Aerobiologia 2007, 23, 211–219. [Google Scholar] [CrossRef]
- Anna, R.D.; Lazzeri, P.; Frisanco, M.; Monti, F.; Campeggi, F.M.; Gottardini, E.; Bersani, M. Pollen discrimination and classification by Fourier transform infrared (FT-IR) microspectroscopy and machine learning. Anal. Bioanal. Chem. 2009, 394, 1443–1452. [Google Scholar]
- Ivleva, N.P.; Niessner, R.; Panne, U. Characterization and discrimination of pollen by Raman microscopy. Anal. Bioanal. Chem. 2005, 381, 261–267. [Google Scholar] [CrossRef]
- Guedes, A.; Ribeiro, H.; Fernández-González, M.; Aira, M.J.; Abreu, I. Pollen Raman spectra database: Application to the identification of airborne pollen. Talanta 2014, 119, 473–478. [Google Scholar] [CrossRef]
- Holt, K.A.; Bennett, K.D. Principles and methods for automated palynology. New Phytol. 2014, 203, 735–742. [Google Scholar] [CrossRef]
- Stillman, E.C.; Flenley, J.R. The needs and prospects for automation in palynology. Quat. Sci. Rev. 1996, 15, 1–5. [Google Scholar] [CrossRef]
- Holt, K.A.; Allen, G.; Hodgson, R.; Marsland, S.; Flenley, J. Progress towards an automated trainable pollen location and classifier system for use in the palynology laboratory. Rev. Palaeobot. Palynol. 2011, 167, 175–183. [Google Scholar] [CrossRef]
- Ticay-Rivas, J.R.; Pozo-Baños, M.; Travieso, C.M.; Arroyo-Hernández, J.; Pérez1, S.T.; Alonso, J.B.; Mora-Mora, F. Pollen Classification Based on Geometrical, Descriptors and Colour Features Using Decorrelation Stretching Method. In Proceedings of the IFIP Advances in Information and Communication Technology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 342–349. [Google Scholar] [Green Version]
- Li, P.; Treloar, W.J.; Flenley, J.R.; Empson, L. Towards automation of palynology 2: The use of texture measures and neural network analysis for automated identification of optical images of pollen grains. J. Quat. Sci. 2004, 19, 755–762. [Google Scholar] [CrossRef]
- Vega, G.L.; Benezeth, Y.; Uhler, M.; Boochs, F.; Vega, G.L.; Benezeth, Y.; Uhler, M.; Boochs, F.; Marzani, F. Sketch of an automatic image based pollen detection system. In Proceedings of the Wissenschaftlich-Technische Jahrestagung der DGPF, Postdam, Germany, 14–16 March 2012; pp. 202–209. [Google Scholar]
- Oteros, J.; Pusch, G.; Weichenmeier, I.; Heimann, U.; Möller, R.; Röseler, S.; Traidl-Hoffmann, C.; Schmidt-Weber, C.; Buters, J.T.M. Automatic and online pollen monitoring. Int. Arch. Allergy Immunol. 2015, 167, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Mularczyk-Oliwa, M.; Bombalska, A.; Kaliszewski, M.; Włodarski, M.; Kopczynski, K.; Kwasny, M.; Szpakowska, M.; Trafny, E.; Zbieta, A. Comparison of fluorescence spectroscopy and FTIR in differentiation of plant pollens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 97, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Pummer, B.G.; Bauer, H.; Bernardi, J.; Chazallon, B.; Facq, S.; Lendl, B.; Whitmorec, K.; Grothe, H. Chemistry and morphology of dried-up pollen suspension residues. J. Raman Spectrosc. 2013, 44, 1654–1658. [Google Scholar] [CrossRef]
- Pappas, C.S.; Tarantilis, P.A.; Harizanis, P.C.; Polissiou, M.G. New Method for Pollen Identification by FT-IR Spectroscopy. Appl. Spectrosc. 2003, 57, 23–27. [Google Scholar] [CrossRef]
- Zimmermann, B. Characterization of Pollen by Vibrational Spectroscopy. Appl. Spectrosc. 2010, 64, 1364–1373. [Google Scholar] [CrossRef]
- Samuels, A.C.; Delucia, F.C.; Mcnesby, K.L.; Miziolek, A.W. Laser-induced breakdown spectroscopy of bacterial spores, molds, pollens, and protein: initial studies of discrimination potential. Appl. Opt. 2003, 42, 6205–6209. [Google Scholar] [CrossRef]
- Boyain-Goitia, A.R.; Beddows, D.C.S.; Griffiths, B.C.; Telle, H.H. Single-pollen analysis by laser-induced breakdown spectroscopy and Raman microscopy. Appl. Opt. 2003, 42, 6119–6132. [Google Scholar] [CrossRef]
- Laucks, M.L.; Roll, G.; Schweiger, G.; Davis, E.J. Physical and Chemical (Raman) Characterization of Bioaerosols—Pollen. J. Aerosol Sci. 2000, 31, 307–319. [Google Scholar] [CrossRef]
- Schulte, F.; Mäder, J.; Kroh, L.W.; Panne, U.; Kneipp, J. Characterization of Pollen Carotenoids with in situ and High-Performance Thin-Layer Chromatography Supported Resonant Raman Spectroscopy. Anal. Chem. 2009, 81, 8426–8433. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M.; Baranski, R. Potential of NIR-FT-Raman Spectroscopy in Natural Carotenoid Analysis. Biopolymers 2005, 77, 212–221. [Google Scholar] [CrossRef]
- Schulte, F.; Lingott, J.; Panne, U.; Kneipp, J. Chemical Characterization and Classification of Pollen. Anal. Chem. 2008, 80, 9551–9556. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S.; Merk, V.; Kneipp, J. Identification of aqueous pollen extracts using surface enhanced Raman scattering (SERS) and pattern recognition methods. J. Biophotonics 2016, 9, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, Y.-L.; Hill, S.C.; Redding, B. Photophoretic Trapping-Raman spectroscopy for single pollens and fungal spores trapped in air. J. Quant. Spectrosc. Radiat. Transf. 2015, 153, 4–12. [Google Scholar] [CrossRef]
- Schulte, F.; Panne, U.; Kneipp, J. Molecular changes during pollen germination can be monitored by Raman microspectroscopy. J. Biophotonics 2010, 3, 542–547. [Google Scholar] [CrossRef]
- Sengupta, A.; Laucks, M.L.; Davis, E.J. Surface-Enhanced Raman Spectroscopy of Bacteria and Pollen. Appl. Spectrosc. 2005, 59, 1016–1023. [Google Scholar] [CrossRef]
- Crouzy, B.; Stella, M.; Konzelmann, T.; Calpini, B.; Clot, B. All-optical automatic pollen identification: Towards an operational system. Atmos. Environ. 2016, 140, 202–212. [Google Scholar] [CrossRef]
- Šauliene, I.; Šukiene, L.; Daunys, G.; Valiulis, G.; Vaitkevičius, L.; Matavulj, P.; Brdar, S.; Panic, M.; Sikoparija, B.; Clot, B.; et al. Automatic pollen recognition with the Rapid-E particle counter: The first-level procedure, experience and next steps. Atmos. Meas. Tech. 2019, 12, 3435–3452. [Google Scholar] [CrossRef]
- Schie, I.W.; Alber, L.; Gryshuk, A.L.; Chan, J.W. Investigating drug induced changes in single, living lymphocytes based on Raman micro-spectroscopy. Analyst 2014, 139, 2726–2733. [Google Scholar] [CrossRef]
- Eberhardt, K.; Beleites, C.; Marthandan, S.; Matthäus, C.; Diekmann, S.; Popp, J. Raman and Infrared Spectroscopy Distinguishing Replicative Senescent from Proliferating Primary Human Fibroblast Cells by Detecting Spectral Differences Mainly Due to Biomolecular Alterations. Anal. Chem. 2017, 89, 2937–2947. [Google Scholar] [CrossRef]
- Notingher, I. Raman spectroscopy cell-based biosensors. Sensors 2007, 7, 1343–1358. [Google Scholar] [CrossRef]
- Schie, I.W.; Rüger, J.; Mondol, A.S.; Ramoji, A.; Neugebauer, U.; Krafft, C.; Popp, J. High-Throughput Screening Raman Spectroscopy Platform for Label-Free Cellomics. Anal. Chem. 2018, 90, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Rüger, J.; Mondol, A.S.; Schie, I.W.; Popp, J.; Krafft, C. High-throughput screening Raman microspectroscopy for assessment of drug-induced changes in diatom cells. Analyst 2019, 144, 4488–4492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhardt, K.; Stiebing, C.; Matthaüs, C.; Schmitt, M.; Popp, J. Advantages and limitations of Raman spectroscopy for molecular diagnostics: An update. Expert Rev. Mol. Diagn. 2015, 15, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Dörfer, T.; Bocklitz, T.; Tarcea, N.; Schmitt, M.; Popp, J. Checking and improving calibration of raman spectra using chemometric approaches. Z. Phys. Chem. 2011, 225, 753–764. [Google Scholar] [CrossRef]
- Martens, H.; Nielsen, J.P.; Engelsen, S.B.S.B. Light scattering and light absorbance separated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 2003, 75, 394–404. [Google Scholar] [CrossRef]
- Cordero, E.; Korinth, F.; Stiebing, C.; Krafft, C.; Schie, I.W.; Popp, J. Evaluation of Shifted Excitation Raman Difference Spectroscopy and Comparison to Computational Background Correction Methods Applied to Biochemical Raman Spectra. Sensors 2017, 17, 1724. [Google Scholar] [CrossRef]
- Revelle, W. Hierarchical Cluster Analysis and the Internal Structure of Tests. Multivariate Behav. Res. 1979, 14, 57–74. [Google Scholar] [CrossRef]
- Singh, A.; Yadav, A.; Rana, A. K-means with Three Different Distance Metrics. Int. J. Comput. Appl. 2013, 67, 13–17. [Google Scholar] [CrossRef]
- Saraçli, S.; Doğan, N.; Doğan, İ. Comparison of hierarchical cluster analysis methods by cophenetic correlation. J. Inequalities Appl. 2013, 2013, 203. [Google Scholar] [CrossRef]
- Shlens, J. A Tutorial on Principal Component Analysis. Syst. Neurobiol. Lab. Univ. Calif. San Diego 2005, 82, 1–12. [Google Scholar]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Support Vector Machines for classification and regression. Analyst 2010, 135, 230–267. [Google Scholar] [CrossRef] [PubMed]
- Brauchle, E.; Thude, S.; Brucker, S.Y.; Schenke-Layland, K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, M.; Schie, I.W.; Tolstik, T.; Stanca, S.E.; Krafft, C.; Popp, J.; Schie, I.W.; Tolstik, T.; Stanca, S.E.; Krafft, C.; et al. Surface-enhanced Raman spectroscopy of cell lysates mixed with silver nanoparticles for tumor classification. Beilstein J. Nanotechnol. 2017, 8, 1183–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassoun, M.; Rüger, J.; Kirchberger-tolstik, T.; Schie, I.W. A droplet-based microfluidic chip as a platform for leukemia cell lysate identification using surface-enhanced Raman scattering. Anal. Bioanal. Chem. 2018, 410, 999–1006. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Van der Maaten, L. Accelerating t-SNE using Tree-Based Algorithms. J. Mach. Learn. Res. 2014, 15, 3221–3245. [Google Scholar]


| Growth Habit | Family | Genus | Growth Habit | Family | Genus |
|---|---|---|---|---|---|
| Grass | Poaceae | Festuca | Shrub | Asteraceae | Artemisia a. |
| Poaceae | Arrhenatherum | Rosaceae | Rosa | ||
| Poaceae | Holcus | Asteraceae | Ambrosia v. | ||
| Poaceae | Phleum | Asteraceae | Helianthus | ||
| Poaceae | Lolium | Asteraceae | Artemisia | ||
| Poaceae | Molinia | ||||
| Poaceae | Elymus | Tree | Betulaceae | Alnus | |
| Poaceae | Calamagrostis | Oleaceae | Fraxinus | ||
| Poaceae | Poa | Ranunculaceae | Ranunculus | ||
| Poaceae | Secale | Betulaceae | Corylus | ||
| Poaceae | Phragmites | Adoxaceae | Sambucus | ||
| Poaceae | Deschampsia | Pinaceae | Larix | ||
| Cyperaceae | Scirpus | Salicaceae | Populus | ||
| Poaceae | Agrostis | Platanaceae | Platanus | ||
| Poaceae | Bromus | Fagaceae | Fagus | ||
| Poaceae | Helictotrichon | Ulmaceae | Ulmus | ||
| Herb | Primulaceae | Cyclamen | Betulaceae | Betula | |
| Polygonaceae | Rumex | ||||
| Amaranthaceae | Amaranthus | ||||
| Papaveraceae | Papaver | ||||
| Plantaginaceae | Plantago |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondol, A.S.; Patel, M.D.; Rüger, J.; Stiebing, C.; Kleiber, A.; Henkel, T.; Popp, J.; Schie, I.W. Application of High-Throughput Screening Raman Spectroscopy (HTS-RS) for Label-Free Identification and Molecular Characterization of Pollen. Sensors 2019, 19, 4428. https://doi.org/10.3390/s19204428
Mondol AS, Patel MD, Rüger J, Stiebing C, Kleiber A, Henkel T, Popp J, Schie IW. Application of High-Throughput Screening Raman Spectroscopy (HTS-RS) for Label-Free Identification and Molecular Characterization of Pollen. Sensors. 2019; 19(20):4428. https://doi.org/10.3390/s19204428
Chicago/Turabian StyleMondol, Abdullah S., Milind D. Patel, Jan Rüger, Clara Stiebing, Andreas Kleiber, Thomas Henkel, Jürgen Popp, and Iwan W. Schie. 2019. "Application of High-Throughput Screening Raman Spectroscopy (HTS-RS) for Label-Free Identification and Molecular Characterization of Pollen" Sensors 19, no. 20: 4428. https://doi.org/10.3390/s19204428




