A Graphene-Based Glycan Biosensor for Electrochemical Label-Free Detection of a Tumor-Associated Antibody
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Electrode Pretreatment
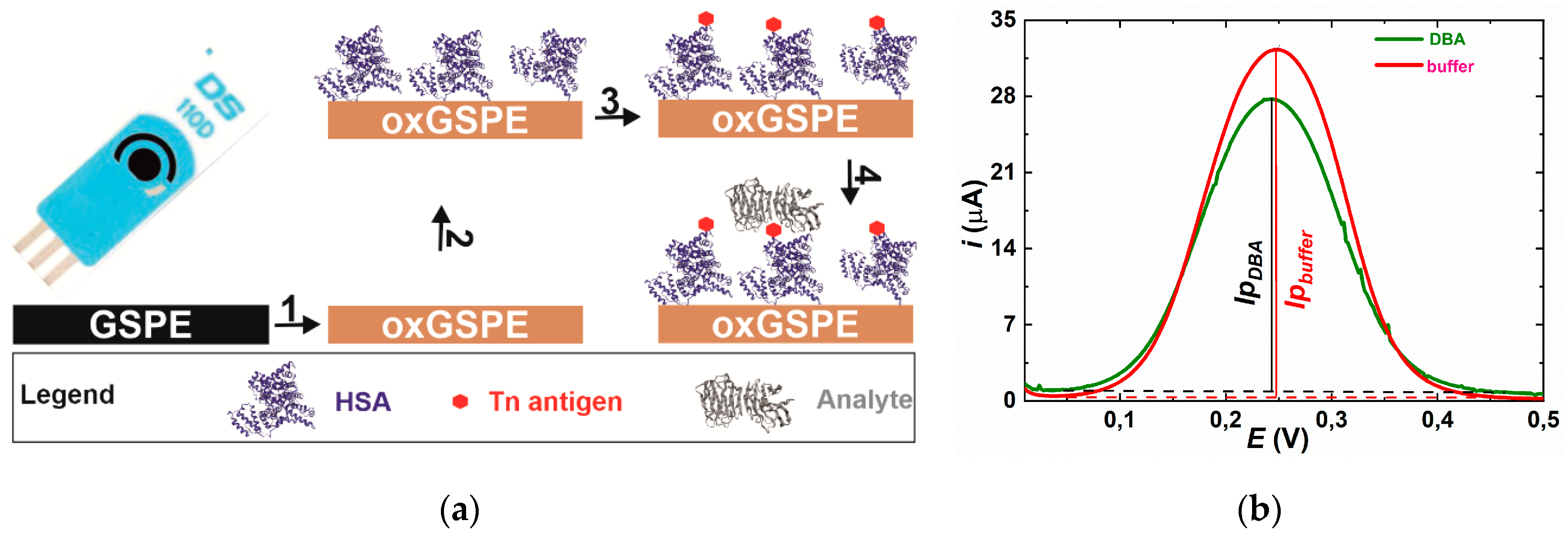
2.3. The Glycan Biosensor
2.4. Differential Pulse Voltammetry (DPV) Measurement
3. Results and Discussion
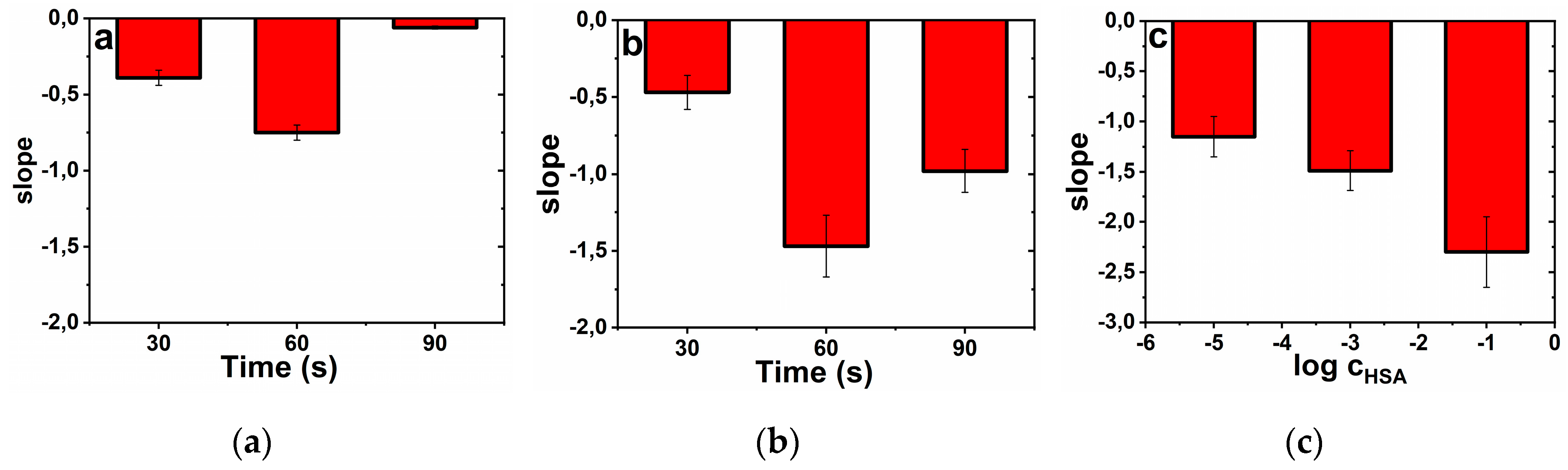
3.1. Optimization of Construction of the Glycan Biosensor
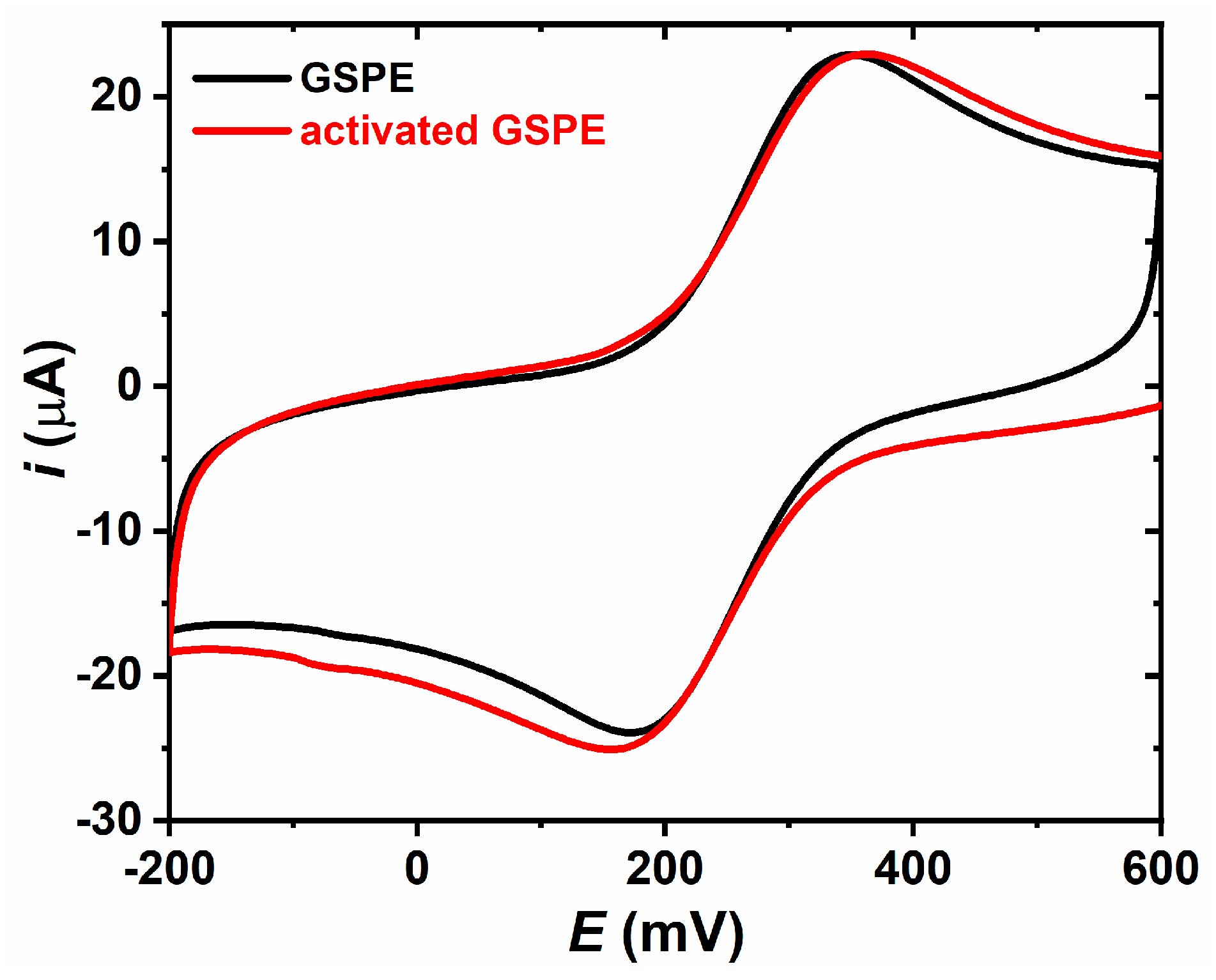
3.1.1. Electrode Activation
3.1.2. Modification of the Electrode by HSA
3.2. Characterization of the Surfaces
3.2.1. Electrochemical Characterization of the Surfaces
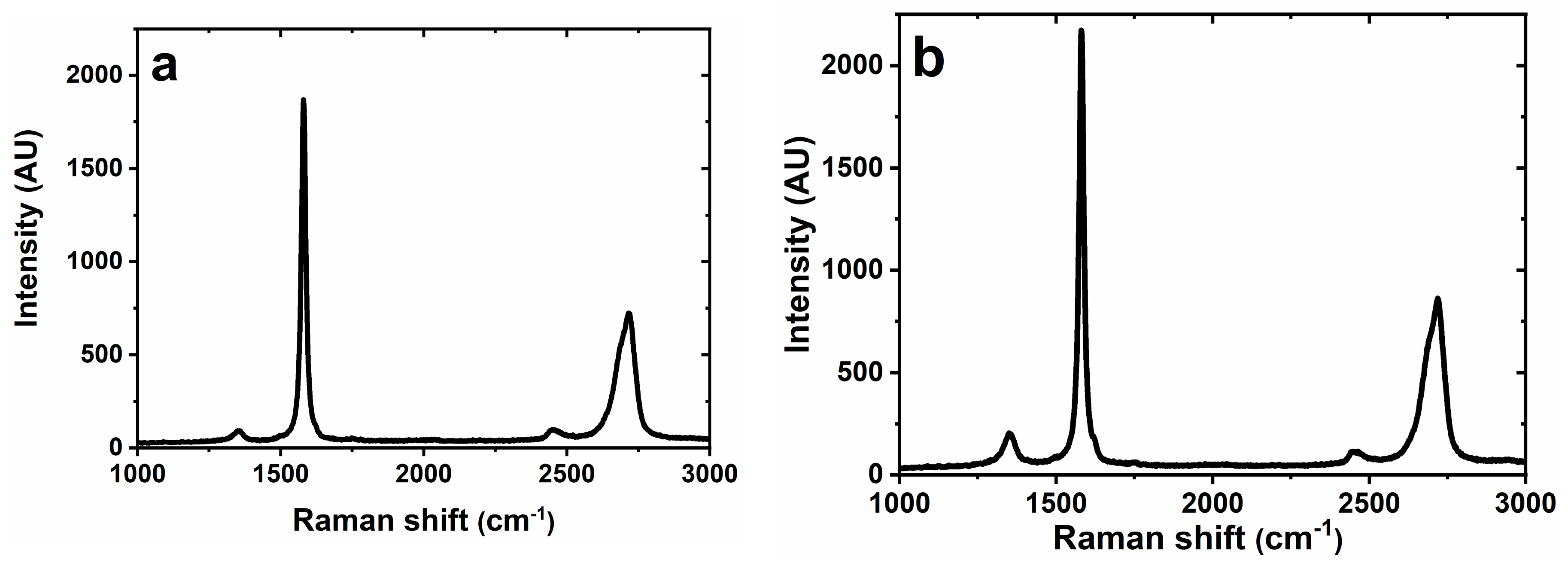
3.2.2. Raman Spectroscopy
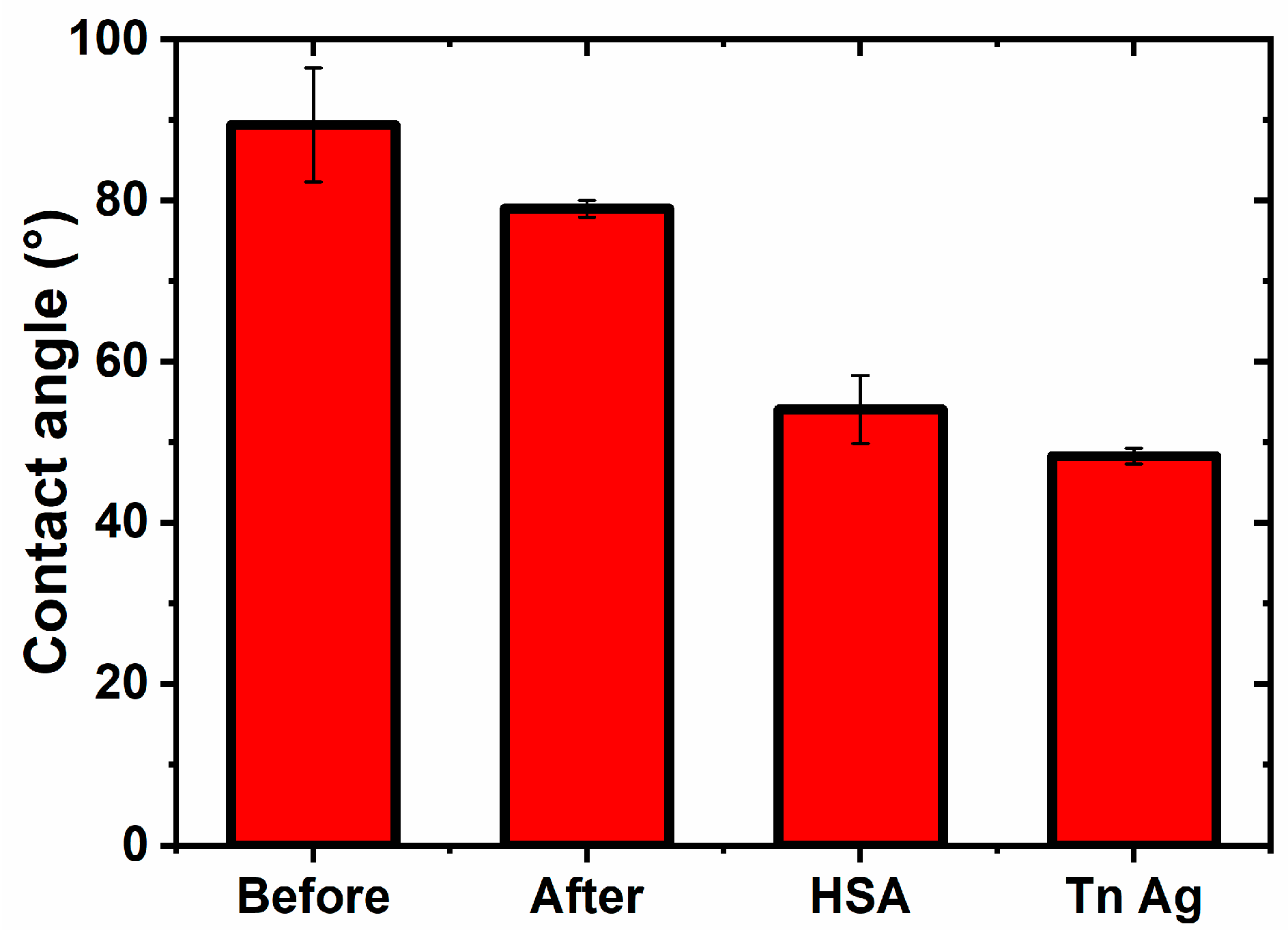
3.2.3. Contact Angle Measurements
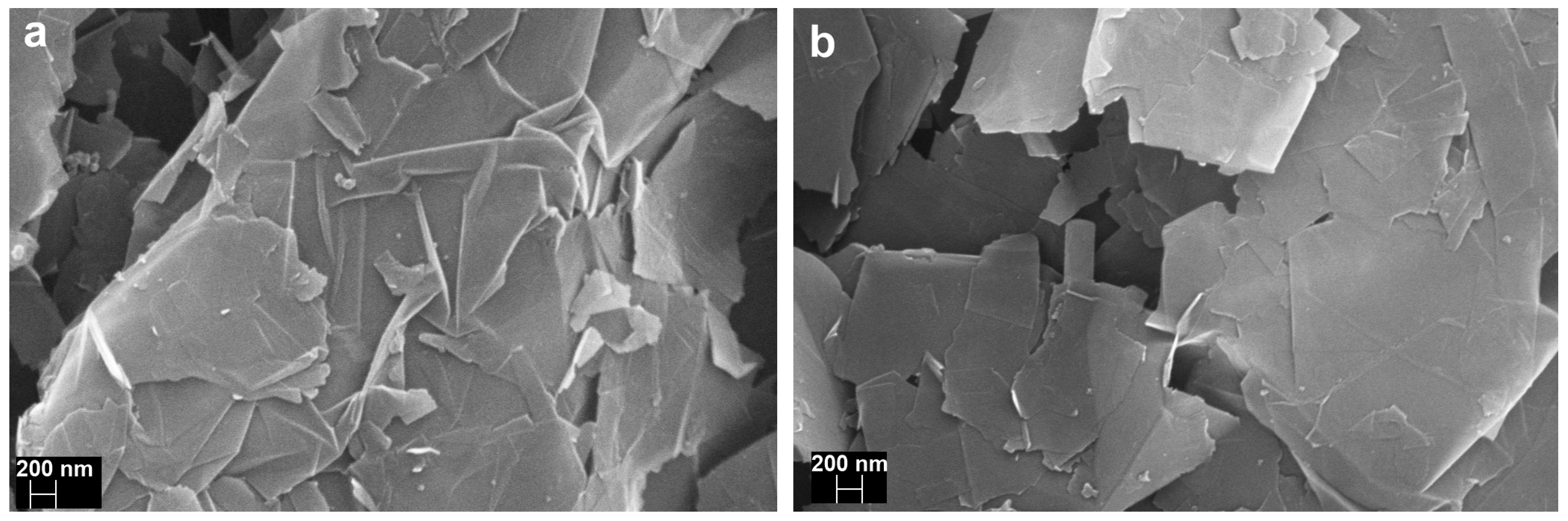
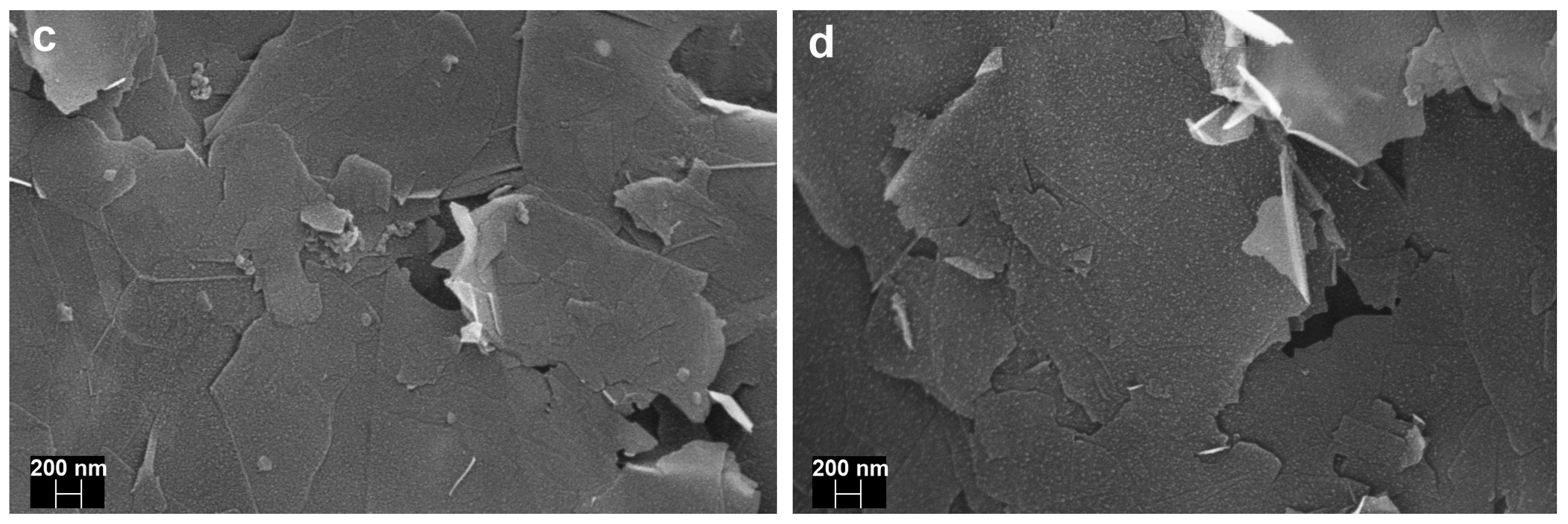
3.2.4. Scanning Electron Microscopy (SEM)
3.2.5. EDX Measurements
3.2.6. Secondary Ion Mass Spectrometry (SIMS) Measurements
3.2.7. Atomic Force Microscopy (AFM) Measurements
3.2.8. The Glycan Biosensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ho, W.L.; Hsu, W.M.; Huang, M.C.; Kadomatsu, K.; Nakagawara, A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J. Hematol. Oncol. 2016, 9, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Liu, M.; Mao, Y.; Xu, Y.; Niu, S. Ultrasensitive photoelectrochemical immunoassay of antibody against tumor-associated carbohydrate antigen amplified by functionalized graphene derivates and enzymatic biocatalytic precipitation. Biosens. Bioelectron. 2014, 59, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics. 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA A Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Springer, G.F. T and Tn, general carcinoma autoantigens. Science 1984, 224, 1198–1206. [Google Scholar] [CrossRef]
- Itzkowitz, S.H.; Yuan, M.; Montgomery, C.K.; Kjeldsen, T.; Takahashi, H.K.; Bigbee, W.L.; Kim, Y.S. Expression of Tn, Sialosyl-Tn, and T Antigens in Human Colon Cancer. Cancer Res. 1989, 49, 197–204. [Google Scholar]
- Richichi, B.; Thomas, B.; Fiore, M.; Bosco, R.; Qureshi, H.; Nativi, C.; Renaudet, O.; BenMohamed, L. A cancer therapeutic vaccine based on clustered Tn-antigen mimetics induces strong antibody-mediated protective immunity. Angew. Chem.-Int. Ed. 2014, 53, 11917–11920. [Google Scholar] [CrossRef]
- Gill, D.J.; Tham, K.M.; Chia, J.; Wang, S.C.; Steentoft, C.; Clausen, H.; Bard-Chapeau, E.A.; Bard, F.A. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc. Natl. Acad. Sci. USA 2013, 110, E3152–E3161. [Google Scholar] [CrossRef] [Green Version]
- Moreau, R.; Dausset, J.; Bernard, J.; Moullec, J. Acquired hemolytic anemia with polyagglutinability of erythrocytes by a new factor present in normal blood. Bull. Mem. Soc. Med. Hop. Paris 1957, 73, 569–587. [Google Scholar]
- Persson, N.; Stuhr-Hansen, N.; Risinger, C.; Mereiter, S.; Polónia, A.; Polom, K.; Kovács, A.; Roviello, F.; Reis, C.A.; Welinder, C.; et al. Epitope mapping of a new anti-Tn antibody detecting gastric cancer cells. Glycobiology 2017, 27, 635–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, W.L.; Chou, C.H.; Jeng, Y.M.; Lu, M.Y.; Yang, Y.L.; Jou, S.T.; Lin, D.T.; Chang, H.H.; Lin, K.H.; Hsu, W.M.; et al. GALNT2 suppresses malignant phenotypes through IGF-1 receptor and predicts favorable prognosis in neuroblastoma. Oncotarget 2014, 5, 12247–12259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganneau, C.; Simenel, C.; Emptas, E.; Courtiol, T.; Coïc, Y.M.; Artaud, C.; Dériaud, E.; Bonhomme, F.; Delepierre, M.; Leclerc, C.; et al. Large-scale synthesis and structural analysis of a synthetic glycopeptide dendrimer as an anti-cancer vaccine candidate. Org. Biomol. Chem. 2017, 15, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56. [Google Scholar] [CrossRef] [PubMed]
- Welinder, C.; Baldetorp, B.; Blixt, O.; Grabau, D.; Jansson, B. Primary Breast Cancer Tumours Contain High Amounts of IgA1 Immunoglobulin: An Immunohistochemical Analysis of a Possible Carrier of the Tumour-Associated Tn Antigen. PLoS ONE 2013, 8, e61749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoja-Łukowicz, D.; Przybyło, M.; Pocheć, E.; Drabik, A.; Silberring, J.; Kremser, M.; Schadendorf, D.; Laidler, P.; Lityńska, A. The new face of nucleolin in human melanoma, Cancer Immunology. Immunotherapy 2009, 58, 1471–1480. [Google Scholar] [CrossRef]
- Qin, Q.; Yin, Z.; Wu, X.; Haas, K.M.; Huang, X. Valency and density matter: Deciphering impacts of immunogen structures on immune responses against a tumor associated carbohydrate antigen using synthetic glycopolymers. Biomaterials 2016, 101, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Zaragoza, M.; Hernández-Ávila, R.; Viedma-Rodríguez, R.; Arenas-Aranda, D.; Ostoa-Saloma, P. Natural and adaptive IgM antibodies in the recognition of tumor-associated antigens of breast cancer (Review). Oncol. Rep. 2015, 34, 1106–1114. [Google Scholar] [CrossRef] [Green Version]
- Tkac, J.; Bertok, T.; Hires, M.; Jane, E.; Lorencova, L.; Kasak, P. Glycomics of prostate cancer: Updates. Expert Rev. Proteomics 2019, 16, 65–76. [Google Scholar] [CrossRef]
- Tkac, J.; Gajdosova, V.; Hroncekova, S.; Bertok, T.; Hires, M.; Jane, E.; Lorencova, L.; Kasak, P. Prostate-specific antigen glycoprofiling as diagnostic and prognostic biomarker of prostate cancer. Interface Focus 2019, 9, 20180077. [Google Scholar] [CrossRef] [Green Version]
- Bertok, T.; Lorencova, L.; Chocholova, E.; Jane, E.; Vikartovska, A.; Kasak, P.; Tkac, J. Electrochemical impedance spectroscopy-based biosensors: Mechanistic principles, analytical examples for assay of protein cancer biomarkers and challenges towards commercialization. ChemElectroChem 2019, 6, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Blsakova, A.; Kveton, F.; Tkac, J. Glycan Modified Interfaces in Biosensing: An Electrochemical Approach. Curr. Opin. Electrochem. 2019, 14, 60–65. [Google Scholar] [CrossRef]
- Paleček, E.; Tkáč, J.; Bartošík, M.; Bertók, T.; Ostatná, V.; Paleček, J. Electrochemistry of Nonconjugated Proteins and Glycoproteins. Toward Sensors for Biomedicine and Glycomics. Chem. Rev. 2015, 115, 2045–2108. [Google Scholar] [CrossRef] [PubMed]
- Dosekova, E.; Filip, J.; Bertok, T.; Both, P.; Kasak, P.; Tkac, J. Nanotechnology in Glycomics: Applications in Diagnostics, Therapy, Imaging, and Separation Processes. Med. Res. Rev. 2017, 37, 514–626. [Google Scholar] [CrossRef]
- Damborska, D.; Bertok, T.; Dosekova, E.; Holazova, A.; Lorencova, L.; Kasak, P.; Tkac, J. Nanomaterial-based biosensors for detection of prostate specific antigen. Microchim. Acta 2017, 184, 3049–3067. [Google Scholar] [CrossRef]
- Wang, S.K.; Cheng, C.M. Glycan-based diagnostic devices: Current progress, challenges and perspectives. Chem. Commun. 2015, 51, 16750–16762. [Google Scholar] [CrossRef]
- Yang, W.; Ratinac, K.R.; Ringer, S.P.; Thordarson, P.; Gooding, J.J.; Braet, F. Carbon Nanomaterials in Biosensors: Should You Use Nanotubes or Graphene? Angew. Chem. Int. Ed. 2010, 49, 2114–2138. [Google Scholar] [CrossRef]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [Green Version]
- Geim, A.K.; Novoselov, K.S. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef]
- Novoselov, K.S.; Fal’ko, V.I.; Colombo, L.; Gellert, P.R.; Schwab, M.G.; Kim, K. A roadmap for graphene. Nature 2012, 490, 192–200. [Google Scholar] [CrossRef]
- Ratinac, K.R.; Yang, W.; Gooding, J.J.; Thordarson, P.; Braet, F. Graphene and Related Materials in Electrochemical Sensing. Electroanalysis 2011, 23, 803–826. [Google Scholar] [CrossRef]
- Klukova, L.; Filip, J.; Belicky, S.; Vikartovska, A.; Tkac, J. Graphene oxide-based electrochemical label-free detection of glycoproteins down to aM level using a lectin biosensor. Analyst 2016, 141, 4278–4282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welinder, C.; Baldetorp, B.; Borrebaeck, C.; Fredlund, B.-M.; Jansson, B. A new murine IgG1 anti-Tn monoclonal antibody with in vivo anti-tumor activity. Glycobiology 2011, 21, 1097–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Pedrero, M.; Sakslund, H.; Hammerich, O.; Pingarron, J. Electrochemical activation of screen-printed carbon strips. Analyst 1996, 121, 345–350. [Google Scholar] [CrossRef]
- Kveton, F.; Blšáková, A.; Hushegyi, A.; Damborsky, P.; Blixt, O.; Jansson, B.; Tkac, J. Optimization of the Small Glycan Presentation for Binding a Tumor-Associated Antibody: Application to the Construction of an Ultrasensitive Glycan Biosensor. Langmuir 2017, 33, 2709–2716. [Google Scholar] [CrossRef] [Green Version]
- Karuwan, C.; Wisitsoraat, A.; Phokharatkul, D.; Sriprachuabwong, C.; Lomas, T.; Nacapricha, D.; Tuantranont, A. A disposable screen printed graphene–carbon paste electrode and its application in electrochemical sensing. RSC Adv. 2013, 3, 25792–25799. [Google Scholar] [CrossRef]
- Randviir, E.P.; Brownson, D.A.; Metters, J.P.; Kadara, R.O.; Banks, C.E. The fabrication, characterisation and electrochemical investigation of screen-printed graphene electrodes. Phys. Chem. Chem. Phys. 2014, 16, 4598–4611. [Google Scholar] [CrossRef] [Green Version]
- Klar, P.; Lidorikis, E.; Eckmann, A.; Verzhbitskiy, I.A.; Ferrari, A.; Casiraghi, C. Raman scattering efficiency of graphene. Physi. Rev. B 2013, 87, 205435. [Google Scholar] [CrossRef] [Green Version]
- Wall, M. The Raman Spectroscopy of Graphene and the Determination of Layer Thickness. Thermo Scientific; Thermo Fisher Scientific Inc.: Madison, WI, USA, 2011. [Google Scholar]
- Raman Spectroscopy Characterization of Graphene & Graphene Oxide. Available online: https://instanano.com/characterization/experimental/raman-graphene.html (accessed on 1 October 2019).
- Vijayarangamuthu, K.; Ahn, S.; Seo, H.; Yoon, S.H.; Park, C.M.; Jeon, K.J. Temporospatial control of graphene wettability. Adv. Mater. 2016, 28, 661–667. [Google Scholar] [CrossRef]
- Chen, Y.N.; Vedala, H.; Kotchey, G.P.; Audfray, A.; Cecioni, S.; Imberty, A.; Vidal, S.; Star, A. Electronic Detection of Lectins Using Carbohydrate-Functionalized Nanostructures: Graphene versus Carbon Nanotubes. Acs Nano 2012, 6, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.W.; Cai, L.; He, X.P.; Chen, G.R.; Long, Y.T. Anthraquinonyl glycoside facilitates the standardization of graphene electrodes for the impedance detection of lectins. Chem. Cent. J. 2014, 8, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.P.; Zhu, B.W.; Zang, Y.; Li, J.; Chen, G.R.; Tian, H.; Long, Y.T. Dynamic tracking of pathogenic receptor expression of live cells using pyrenyl glycoanthraquinone-decorated graphene electrodes. Chem. Sci. 2015, 6, 1996–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Electrode | Glycan | Immobilization | Detection | Analyte | LOD | Ref. |
|---|---|---|---|---|---|---|
| CCG | pyrene-functionalized monosaccharides | π–π stacking interactions | FET sensing | Lectins | ~1 nM | [42] |
| graphene | antraquinonyl-modified monosaccharides | π–π stacking interactions | DPV | Lectins | ~1 µM | [43] |
| graphene SPE | pyrenyl-anthraquinone monosaccharides | π–π stacking interactions | DPV | Lectins | ~50 nM | [44] |
| graphene SPE | pyrenyl-anthraquinone monosaccharides | π–π stacking interactions | DPV | Cells | ~200-400 cells/mL | [44] |
| oxidized graphene | Tn antigen | covalent on HSA | DPV | Lectin (DBA) | ~1 aM | This work |
| oxidized graphene | Tn antigen | covalent on HSA | DPV | Antibody (2C4) | ~10 aM | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kveton, F.; Blsakova, A.; Lorencova, L.; Jerigova, M.; Velic, D.; Blixt, O.; Jansson, B.; Kasak, P.; Tkac, J. A Graphene-Based Glycan Biosensor for Electrochemical Label-Free Detection of a Tumor-Associated Antibody. Sensors 2019, 19, 5409. https://doi.org/10.3390/s19245409
Kveton F, Blsakova A, Lorencova L, Jerigova M, Velic D, Blixt O, Jansson B, Kasak P, Tkac J. A Graphene-Based Glycan Biosensor for Electrochemical Label-Free Detection of a Tumor-Associated Antibody. Sensors. 2019; 19(24):5409. https://doi.org/10.3390/s19245409
Chicago/Turabian StyleKveton, Filip, Anna Blsakova, Lenka Lorencova, Monika Jerigova, Dusan Velic, Ola Blixt, Bo Jansson, Peter Kasak, and Jan Tkac. 2019. "A Graphene-Based Glycan Biosensor for Electrochemical Label-Free Detection of a Tumor-Associated Antibody" Sensors 19, no. 24: 5409. https://doi.org/10.3390/s19245409





