A Method for Broccoli Seedling Recognition in Natural Environment Based on Binocular Stereo Vision and Gaussian Mixture Model
Abstract
:1. Introduction
2. Materials and Methods
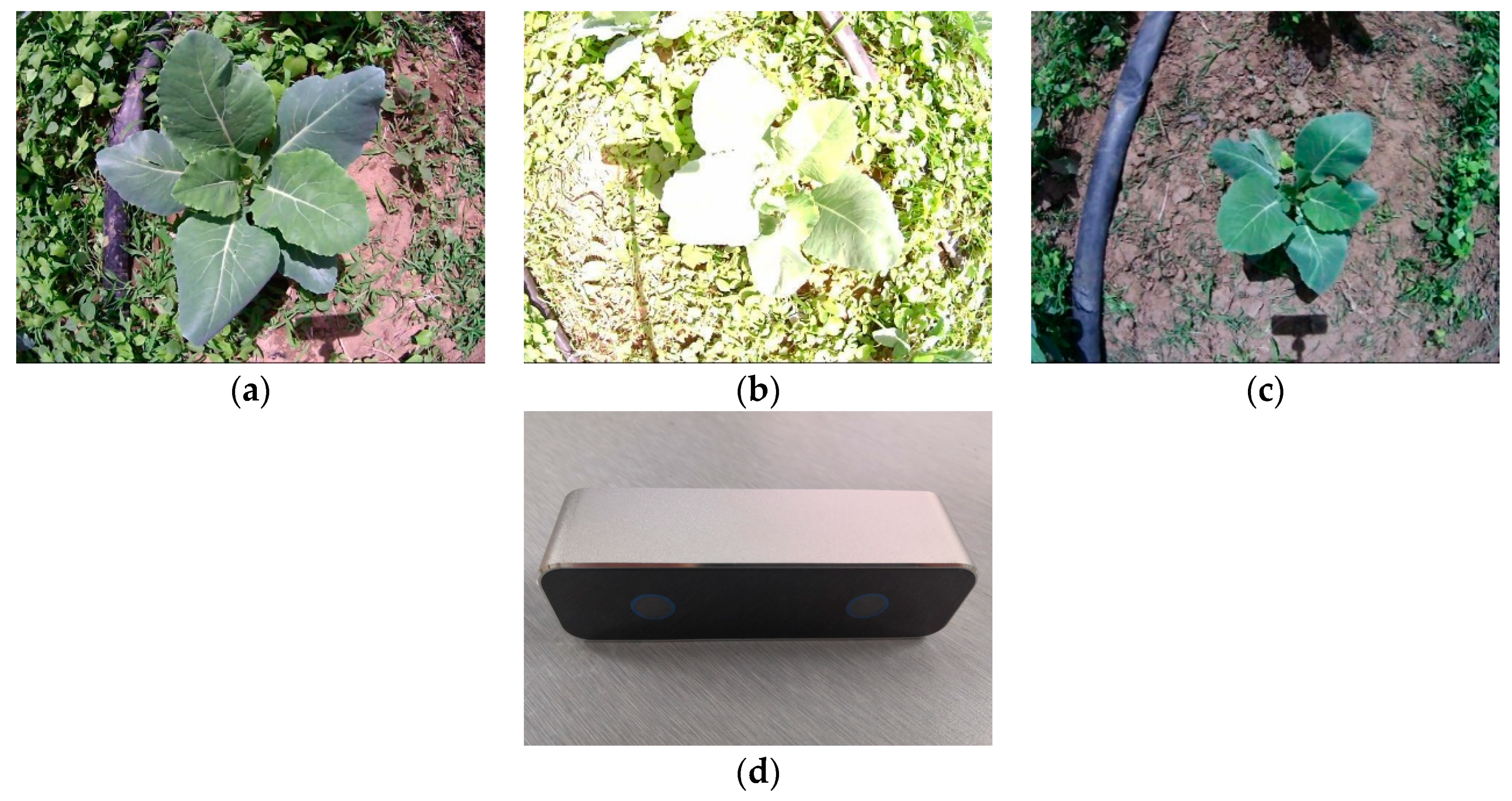
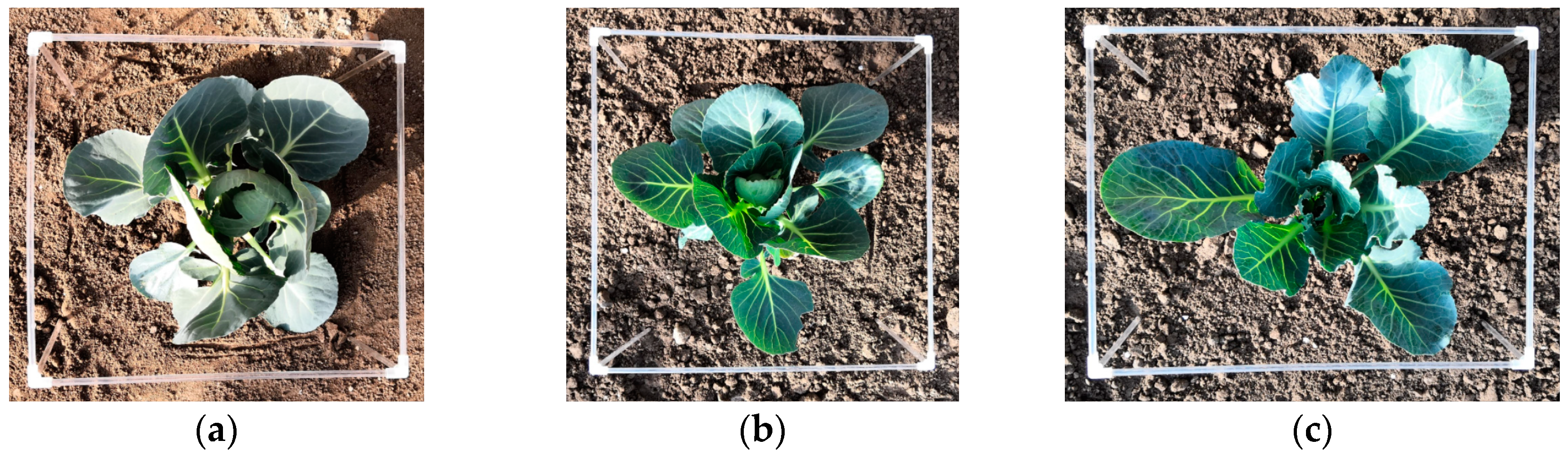
2.1. Image Acquisition and Experiment Platform
2.2. Methods
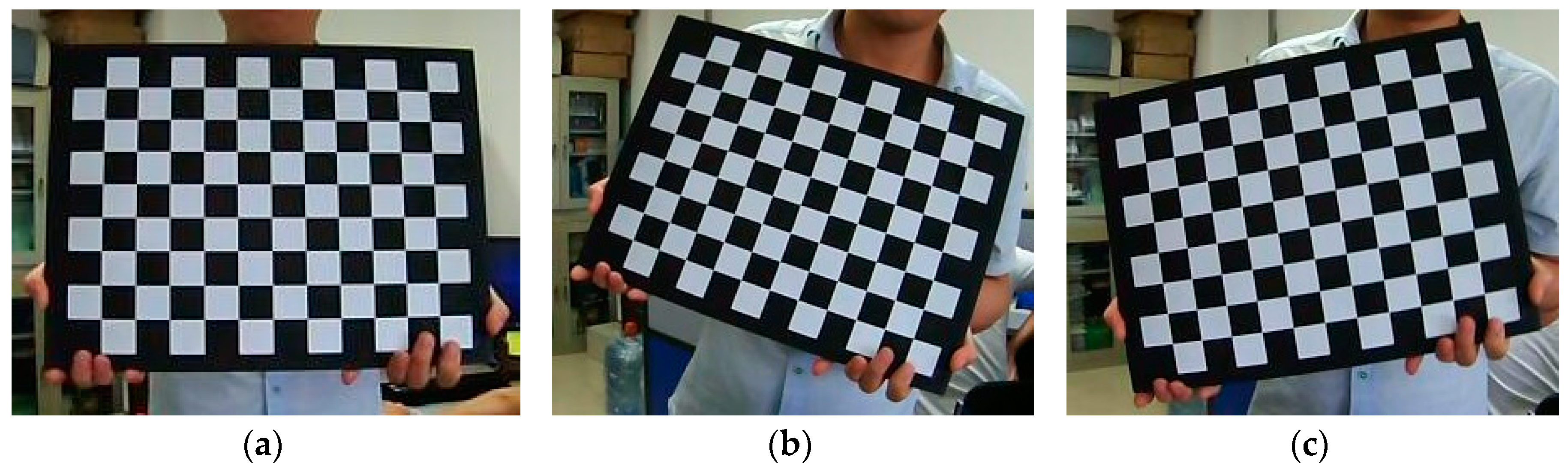
2.2.1. Binocular Camera Calibration
2.2.2. Stereo Rectification
2.2.3. Semi-Global Matching Algorithm and 3D Recognition
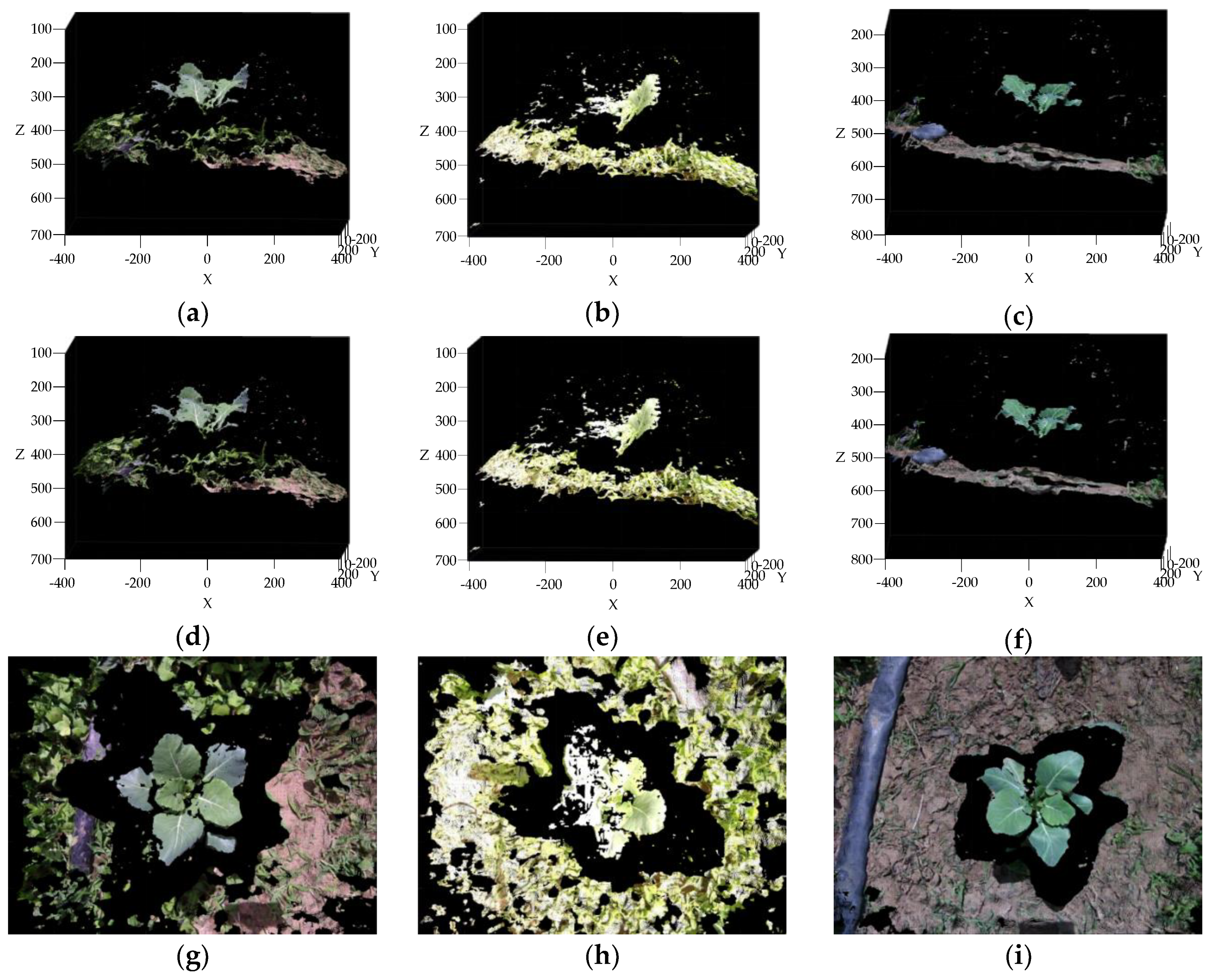
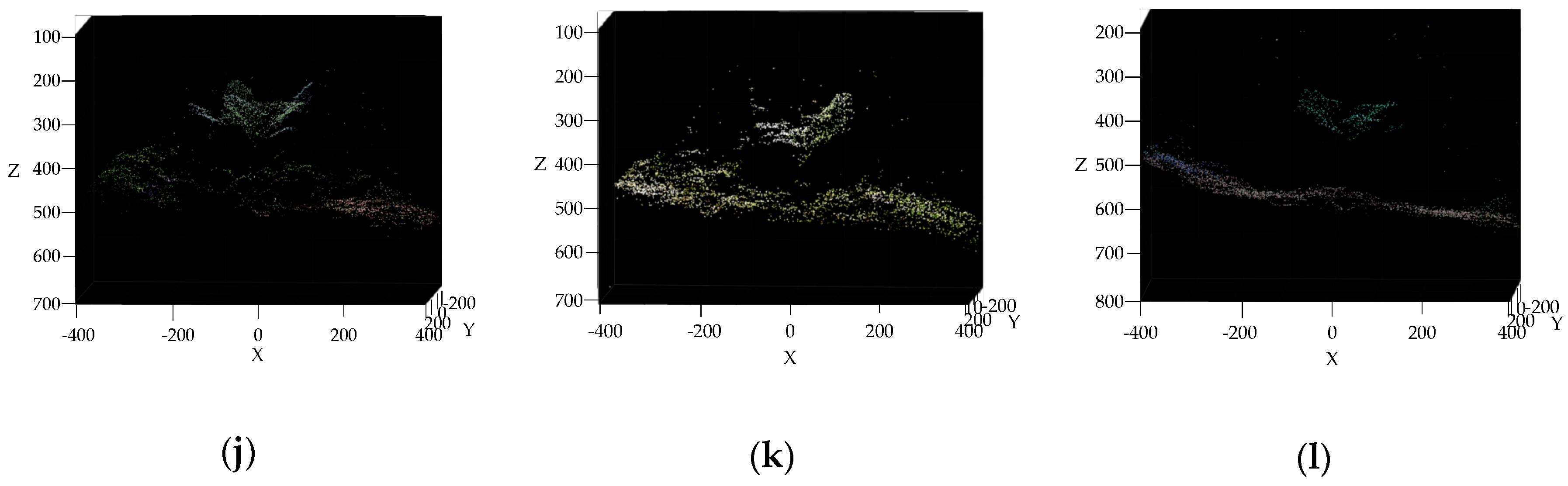
2.2.4. Invalid Points Removal and Down-Sampling
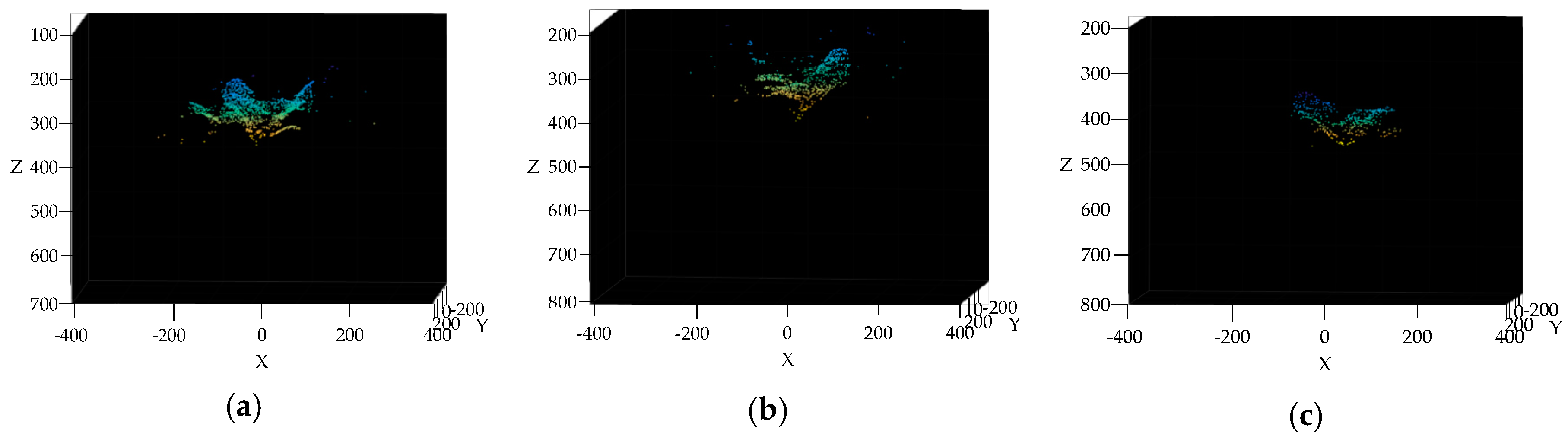
2.2.5. Gaussian Mixture Model cluster
- (1)
- Let be the number of the cluster of the broccoli seedling point cloud, and set initial values of , , separately.
- (2)
- Calculate the posterior probability by using Equation (2) according to the current , , .
- (3)
- Calculate the new , , by using the Equations (3–5).
- (4)
- Calculate the logarithmic likelihood function of Equation (1).
- (5)
- Check whether the parameters , , are convergent or the function (6) is convergent, if not return to (2).
- (6)
- If converge, calculate posterior probability of each point of broccoli seedling point cloud separately, and then categorize the point to the cluster, where has the maximum value.
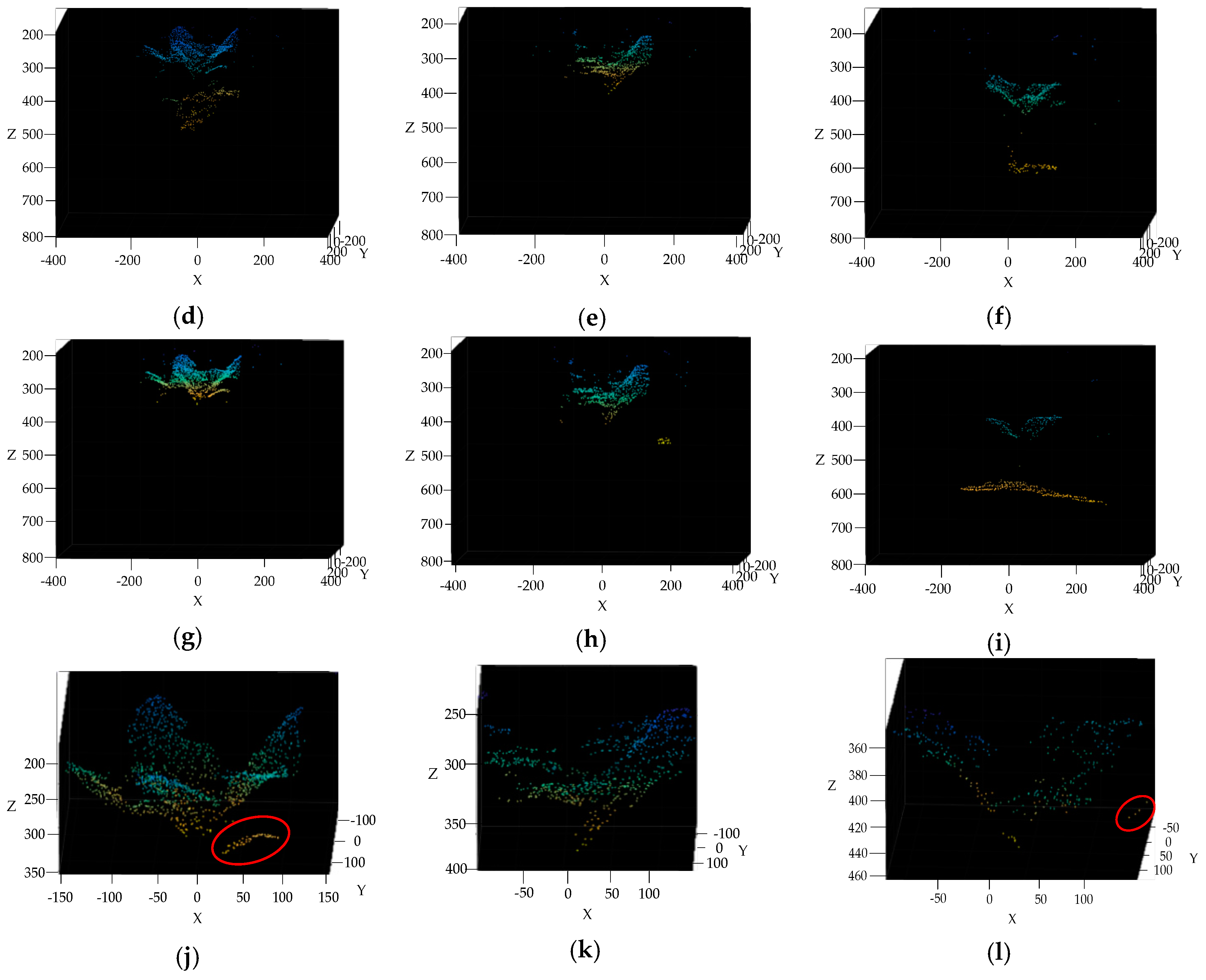
2.2.6. Outlier Filtering by K-Nearest Neighbors (KNN) Algorithm
3. Results
3.1. Stereo Rectification Analysis
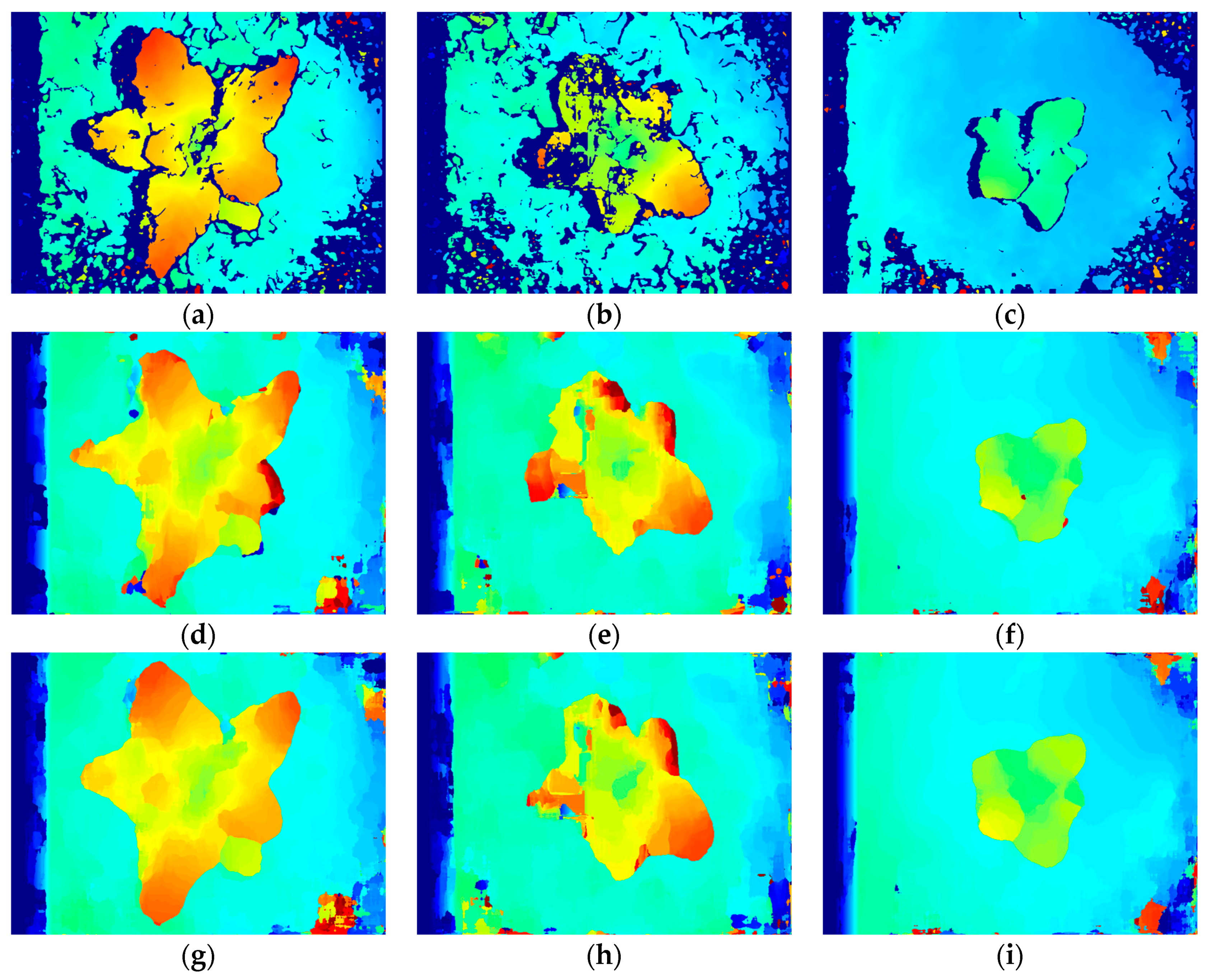
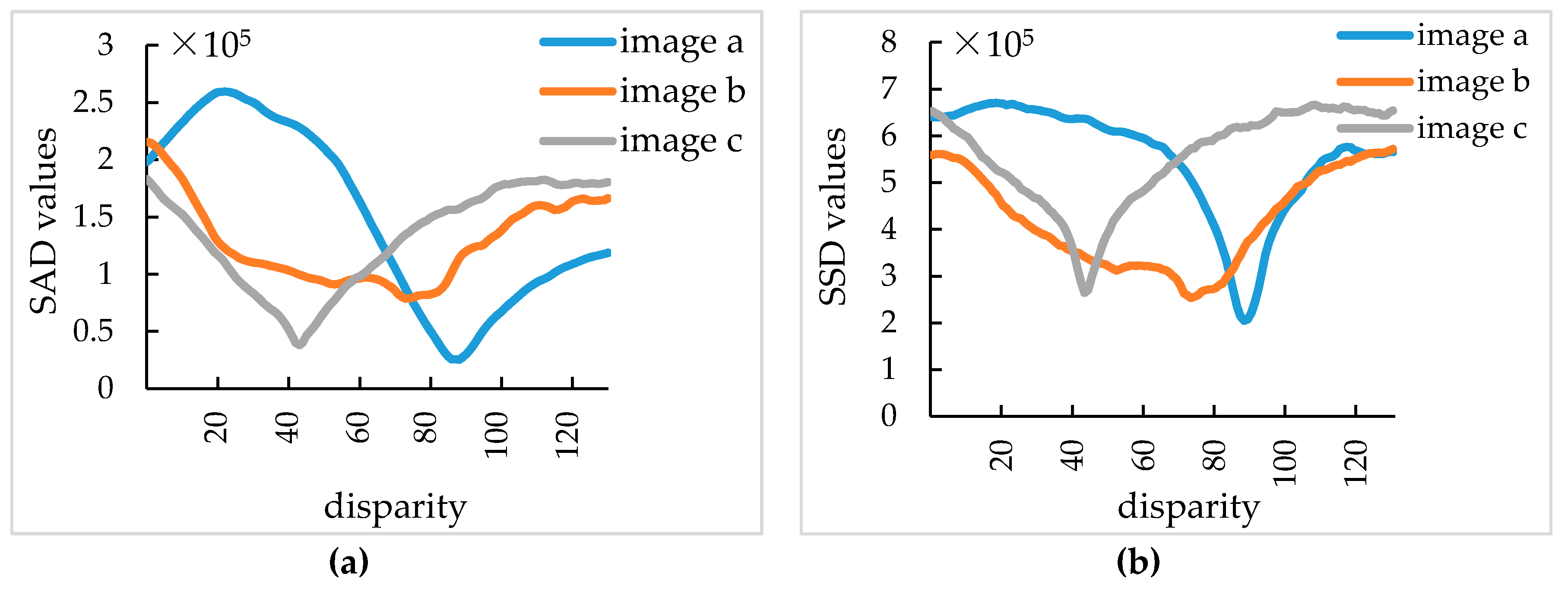
3.2. Stereo Matching Results Analysis
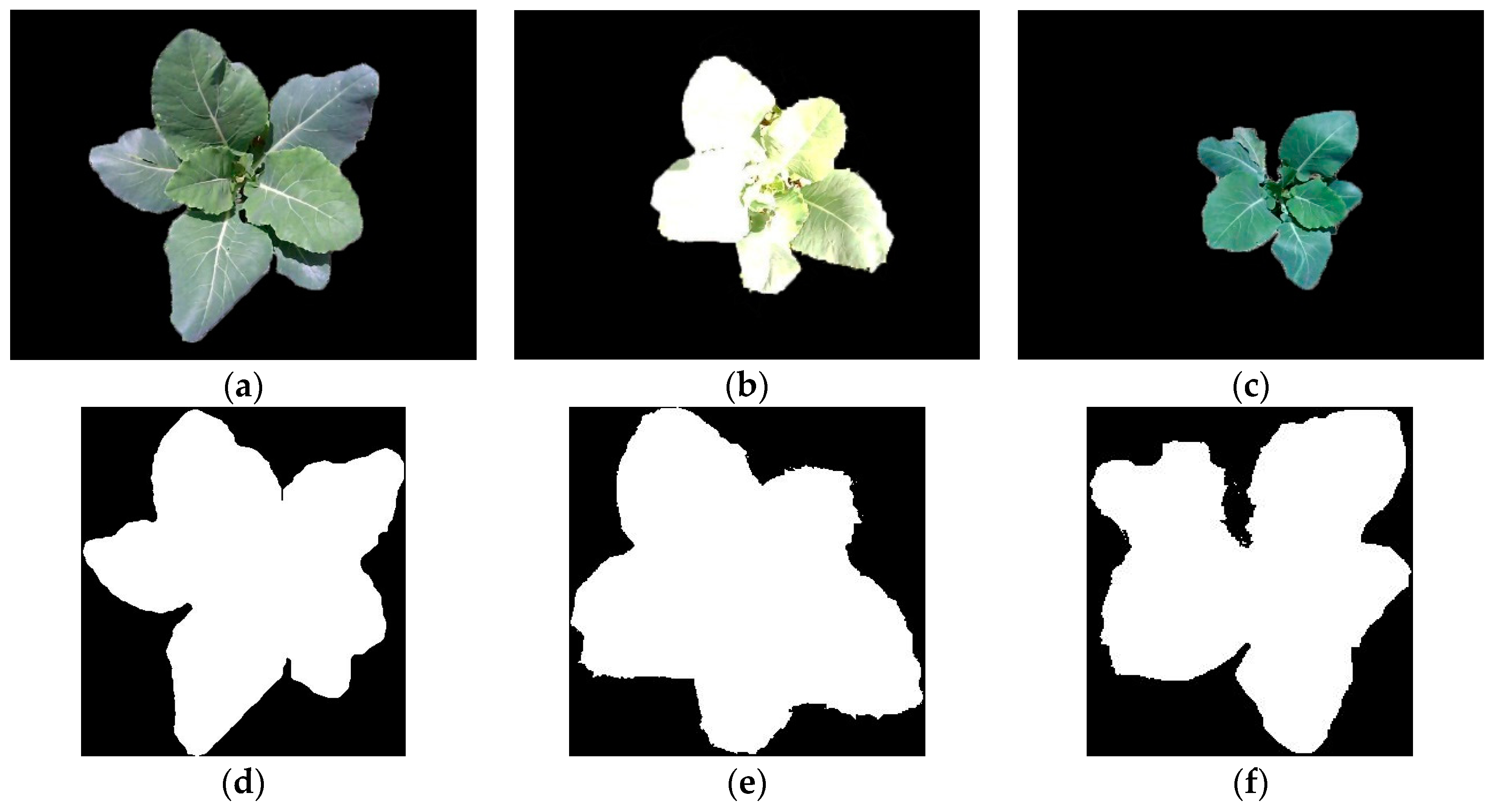
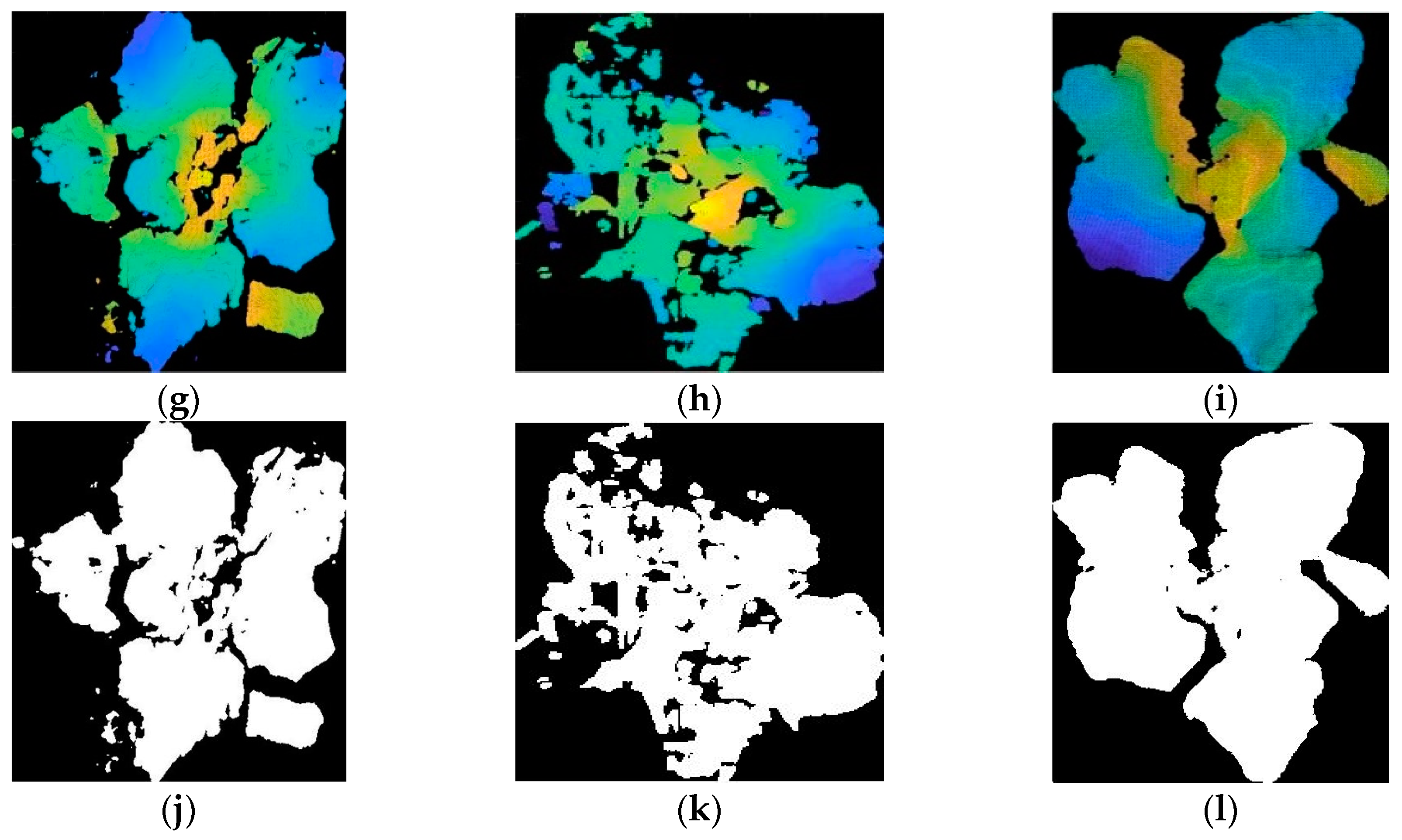
3.3. Reconstruction, Invalid Points Removal and Down-Sampling Results Analysis
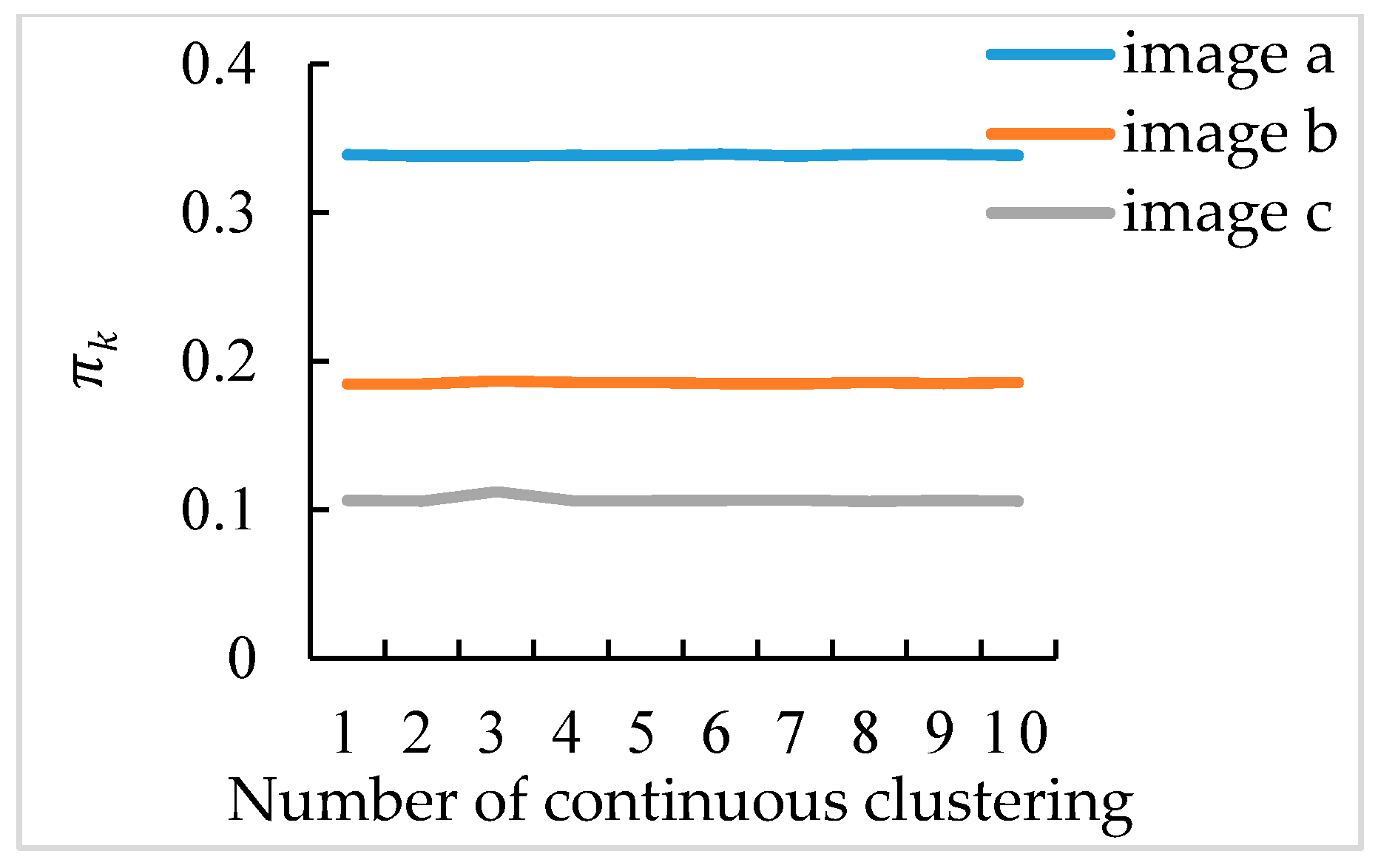
3.4. Broccoli Seedling Points Clustering and Recognition Results Analysis
3.5. Completeness of Broccoli Seedling Recognition
3.6. Measured and Theoretical Envelope Box Volumes
4. Conclusions
- (1)
- A method of broccoli seedling recognition was proposed in this paper, which is based on Binocular Stereo Vision and Gaussian Mixture Model clustering, under different weed conditions, different shooting heights, and different exposure intensities in a natural field. The method was proposed for the rapid identification of transplanted broccoli seedlings with growth advantage. The experimental results of 247 pairs of images proved that correct recognition rate of this method is 97.98%, and the average operation time to process a pair of original images with the resolution of 640×480 was 578 ms. The average value of sensitivity is 85.91%. For cabbage planta the average percentage of the theoretical envelope box volume to the measured envelope box volume is 95.66%.
- (2)
- The SGM algorithm was introduced for a pair of broccoli seedling images with the resolution of 791×547 after stereo rectification. The SGM algorithm was compared with the SAD algorithm and the SSD algorithm. The SGM algorithm can meet the matching requirements of all broccoli seedling images, when the matching window size was 15×15 pixel and the maximum disparity was 128 pixel. The operation time of SGM algorithm was 138 ms. The experimental results showed that SGM algorithm is superior to SAD algorithm and SSD algorithm.
- (3)
- The GMM cluster was adopted for recognizing broccoli seedling points rapidly and stably. The experimental results showed that the proposed GMM algorithm was better than the K-means algorithm and the fuzzy c-means algorithm on recognition effect and stability. The average calculation time of the GMM algorithm was only 51 ms which satisfied the real-time requirements. The KNN algorithm was used for outliers filtering of broccoli seedling points recognized by GMM cluster, and complete and pure broccoli seedling was recognized finally.
Author Contributions
Funding
Conflicts of Interest
References
- Wang, C.; Tang, Y.; Zou, X.; Luo, L.; Chen, X. Recognition and Matching of Clustered Mature Litchi Fruits Using Binocular Charge-Coupled Device (CCD) Color Cameras. Sensors 2017, 17, 2564. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, M.; Zhang, X. 3D Reconstruction of Orchid Based on Virtual Binocular Vision Technology. In Proceedings of the International Conference on Information Science and Control Engineering, Changsha, Hunan, China, 21–23 July 2017; pp. 1–5. [Google Scholar]
- Stein, M.; Bargoti, S.; Underwood, J. Image based mango fruit detection, localisation and yield estimation using multiple view geometry. Sensors 2016, 16, 1915. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, A.L. Improving depth maps of plants by using a set of five cameras. J. Electron. Imaging 2015, 24. [Google Scholar] [CrossRef]
- Wang, Z.; Walsh, K.B.; Verma, B. On-Tree Mango Fruit Size Estimation Using RGB-D Images. Sensors 2017, 17, 2738. [Google Scholar] [CrossRef] [PubMed]
- Andujar, D.; Dorado, J.; Fernandez-Quintanilla, C.; Ribeiro, A. An approach to the use of depth cameras for weed volume estimation. Sensors 2016, 16, 972. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Cheng, W.; Zhou, J.; Wang, X. Design of structured-light vision system for tomato harvesting robot. Int. J. Agric. Biol. Eng. 2014, 7, 19–26. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.H.; Chahl, J.S. Registration of multispectral 3D points for plant inspection. Precis. Agric. 2018, 19, 513–536. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.H.; Chahl, J.S. A multispectral 3D vision system for invertebrate detection on crops. IEEE Sens. J. 2017, 17, 7502–7515. [Google Scholar] [CrossRef]
- Wahabzada, M.; Paulus, S.; Kersting, K.; Mahlein, A.K. Automated interpretation of 3D laserscanned point clouds for plant organ segmentation. BMC Bioinform. 2015, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.; Barron, J.L. Machine Vision System for 3D Plant Phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinf. 2018. [Google Scholar] [CrossRef] [PubMed]
- Barnea, E.; Mairon, R.; Ben-Shahar, O. Colour-agnostic shape-based 3D fruit detection for crop harvesting robots. Biosyst. Eng. 2016, 146, 57–70. [Google Scholar] [CrossRef]
- Si, Y.; Liu, G.; Feng, J. Location of apples in trees using stereoscopic vision. Comput. Electron. Agric. 2015, 112, 68–74. [Google Scholar] [CrossRef]
- Ji, W.; Meng, X.; Qian, Z.; Xu, B.; Zhao, D. Branch localization method based on the skeleton feature extraction and stereo matching for apple harvesting robot. Int. J. Adv. Robot. Syst. 2017, 14, 1–9. [Google Scholar] [CrossRef]
- Gong, L.; Chen, R.; Zhao, Y.; Liu, C. Model-based in-situ measurement of pakchoi leaf area. Int. J. Agric. Biol. Eng. 2015, 8, 35–42. [Google Scholar] [CrossRef]
- Polder, G.; Hofstee, J.W. Phenotyping large tomato plants in the greenhouse using a 3D light-field camera. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting 2014, Montreal, QC, Canada, 13–16 July 2014; pp. 153–159. [Google Scholar]
- Li, J.; Tang, L. Crop recognition under weedy conditions based on 3D imaging for robotic weed control. J. Field Robot. 2018, 35, 596–611. [Google Scholar] [CrossRef]
- Andujar, D.; Ribeiro, A.; Fernandez-Quintanilla, C.; Dorado, J. Using depth cameras to extract structural parameters to assess the growth state and yield of cauliflower crops. Comput. Electron. Agric. 2016, 122, 67–73. [Google Scholar] [CrossRef]
- Avendano, J.; Ramos, P.J.; Prieto, F.A. A System for Classifying Vegetative Structures on Coffee Branches based on Videos Recorded in the Field by a Mobile Device. Expert Syst. Appl. 2017, 88, 178–192. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Vandevoorde, K.; Wouters, N.; Kayacan, E.; De Baerdemaeker, J.G.; Saeys, W. Detection of red and bicoloured apples on tree with an RGB-D camera. Biosyst. Eng. 2016, 146, 33–44. [Google Scholar] [CrossRef]
- Jafari, A.; Bakhshipour, A. A novel algorithm to recognize and locate pomegranate on the tree for the harvesting robot using stereo vision system. In Proceedings of the Precision Agriculture 2011—Papers Presented at the 8th European Conference on Precision Agriculture 2011, Prague, Czech Republic, 11–14 July 2011; pp. 133–142. [Google Scholar]
- Wang, C.; Zou, X.; Tang, Y.; Luo, L.; Feng, W. Localisation of litchi in an unstructured environment using binocular stereo vision. Biosyst. Eng. 2016, 145, 39–51. [Google Scholar] [CrossRef]
- Bac, C.W.; Hemming, J.; Van Henten, E.J. Stem localization of sweet-pepper plants using the support wire as a visual cue. Comput. Electron. Agric. 2014, 105, 111–120. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, X.; Tang, L.; Che, J.; Sun, Y.; Chen, J. Intra-row weed recognition using plant spacing information in stereo images. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting 2013, Kansas City, MO, USA, 21–24 July 2013; pp. 915–921. [Google Scholar]
- Vazquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]
- Mehta, S.S.; Ton, C.; Asundi, S.; Burks, T.F. Multiple camera fruit localization using a particle filter. Comput. Electron. Agric. 2017, 142, 139–154. [Google Scholar] [CrossRef]
- Fernandez, R.; Salinas, C.; Montes, H.; Sarria, J. Multisensory system for fruit harvesting robots. Experimental testing in natural scenarios and with different kinds of crops. Sensors 2014, 14, 23885–23904. [Google Scholar] [CrossRef] [PubMed]
- Shafiekhani, A.; Kadam, S.; Fritschi, F.B.; DeSouza, G.N. Vinobot and Vinoculer: Two Robotic Platforms for High-Throughput Field Phenotyping. Sensors 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Moeckel, T.; Dayananda, S.; Nidamanuri, R.R.; Nautiyal, S.; Hanumaiah, N.; Buerkert, A.; Wachendorf, M. Estimation of Vegetable Crop Parameter by Multi-temporal UAV-Borne Images. Remote Sens. 2018, 10. [Google Scholar] [CrossRef]
- Karpina, M.; Jarząbek-Rychard, M.; Tymkw, P.; Borkowski, A. UAV-based automatic tree growth measurement for biomass estimation. In Proceedings of the 23rd International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences Congress, Prague, Czech Republic, 12–19 July 2016; pp. 685–688. [Google Scholar]
- Lu, H.; Tang, L.; Whitham, S.A. Development of an automatic maize seedling phenotyping platfrom using 3D vision and industrial robot arm. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting 2015, New Orleans, LA, USA, 26–29 July 2015; pp. 4001–4013. [Google Scholar]
- Paulus, S.; Dupuis, J.; Mahlein, A.K.; Kuhlmann, H. Surface feature based classification of plant organs from 3D laserscanned point clouds for plant phenotyping. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Burks, T.F.; Lee, W.S. 3D reconstruction of small plant from multiple views. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting 2014, Montreal, QC, Canada, 13–16 July 2014; pp. 661–668. [Google Scholar]
- Yeh, Y.H.F.; Lai, T.C.; Liu, T.Y.; Liu, C.C.; Chung, W.C.; Lin, T.T. An automated growth measurement system for leafy vegetables. In Proceedings of the 4th International Workshop on Computer Image Analysis in Agriculture, Valencia, Spain, 8–12 July 2012; pp. 43–50. [Google Scholar]
- Zhang, Y.; Teng, P.; Shimizu, Y.; Hosoi, F.; Omasa, K. Estimating 3D leaf and stem shape of nursery paprika plants by a novel multi-camera photography system. Sensors 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.C.; Kicherer, A.; Wieland, M.; Klingbeil, L.; Topfer, R.; Kuhlmann, H. Towards automated large-scale 3D phenotyping of vineyards under field conditions. Sensors 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Guo, X.; Wang, Y.; Zhao, C.; Liao, W. Constructing a Three-Dimensional Resource Database of Plants Using Measured in situ Morphological Data. Appl. Eng. Agric. 2017, 33, 747–756. [Google Scholar] [CrossRef]
- Hui, F.; Zhu, J.; Hu, P.; Meng, L.; Zhu, B.; Guo, Y.; Li, B.; Ma, Y. Image-based dynamic quantification and high-accuracy 3D evaluation of canopy structure of plant populations. Ann. Bot. 2018, 121, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, L. Developing a low-cost 3D plant morphological traits characterization system. Comput. Electron. Agric. 2017, 143, 1–13. [Google Scholar] [CrossRef]
- An, N.; Welch, S.M.; Markelz, R.J.C.; Baker, R.L.; Palmer, C.M.; Ta, J.; Maloof, J.N.; Weinig, C. Quantifying time-series of leaf morphology using 2D and 3D photogrammetry methods for high-throughput plant phenotyping. Comput. Electron. Agric. 2017, 135, 222–232. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, L. Field-based Robotic Phenotyping for Sorghum Biomass Yield Component Traits Characterization Using Stereo Vision. In Proceedings of the 5th IFAC Conference on Sensing, Control and Automation Technologies for Agriculture, Seattle, WA, USA, 14–17 August 2016; pp. 265–270. [Google Scholar]
- Bao, Y.; Tang, L.; Schnable, P.S.; Salas-Fernandez, M.G. Infield Biomass Sorghum Yield Component Traits Extraction Pipeline Using Stereo Vision. In Proceedings of the 2016 ASABE Annual International Meeting, Orlando, FL, USA, 17–20 July 2016. [Google Scholar]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Dupuis, J.; Kuhlmann, H.; Oerke, E.C.; Plumer, L. Generation and application of hyperspectral 3D plant models: methods and challenges. Mach. Vis. Appl. 2016, 27, 611–624. [Google Scholar] [CrossRef]
- Golbach, F.; Kootstra, G.; Damjanovic, S.; Otten, G.; van de Zedde, R. Validation of plant part measurements using a 3D reconstruction method suitable for high-throughput seedling phenotyping. Mach. Vis. Appl. 2016, 27, 663–680. [Google Scholar] [CrossRef]
- Santos, T.T.; Koenigkan, L.V.; Barbedo, J.G.A.; Rodrigues, G.C. 3D plant modeling: localization, mapping and segmentation for plant phenotyping using a single hand-held camera. In Proceedings of the 13th European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 247–263. [Google Scholar]
- Rist, F.; Herzog, K.; Mack, J.; Richter, R.; Steinhage, V.; Topfer, R. High-Precision Phenotyping of Grape Bunch Architecture Using Fast 3D Sensor and Automation. Sensors 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Moriondo, M.; Leolini, L.; Stagliano, N.; Argenti, G.; Trombi, G.; Brilli, L.; Dibari, C.; Leolini, C.; Bindi, M. Use of digital images to disclose canopy architecture in olive tree. Sci. Hortic. 2016, 209, 1–13. [Google Scholar] [CrossRef]
- Duan, T.; Chapman, S.C.; Holland, E.; Rebetzke, G.J.; Guo, Y.; Zheng, B. Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J. Exp. Bot. 2016, 67, 4523–4534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaivivatrakul, S.; Tang, L.; Dailey, M.N.; Nakarmi, A.D. Automatic morphological trait characterization for corn plants via 3D holographic reconstruction. Comput. Electron. Agric. 2014, 109, 109–123. [Google Scholar] [CrossRef]
- Rose, J.C.; Paulus, S.; Kuhlmann, H. Accuracy analysis of a multi-view stereo approach for phenotyping of tomato plants at the organ level. Sensors 2015, 15, 9651–9665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. A flexible new technique for camera calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef] [Green Version]
- Tsai, R.Y. A versatile camera calibration technique for high-accuracy 3D machine vision metrology using off-the-shelf TV cameras and lenses. IEEE J. Rob. Autom. 1987, 3, 323–344. [Google Scholar] [CrossRef]
- Hirschmuller, H. Accurate and Efficient Stereo Processing by Semi-Global Matching and Mutual Information. In Proceedings of the Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; pp. 807–814. [Google Scholar]
- Pomerleau, F.; Colas, F.; Siegwart, R.; Magnenat, S. Comparing ICP variants on real-world data sets. Auton. Robot. 2013, 34, 133–148. [Google Scholar] [CrossRef]
- McLachlan, G.; Peel, D. Finite Mixture Models; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2000. [Google Scholar]
- Rusu, R.B.; Marton, Z.C.; Blodow, N.; Dolha, M.; Beetz, M. Towards 3D Point cloud based object maps for household environments. In Proceedings of the IEEE International Conference on Robotics and Automation, Rome, Italy, 10–14 April 2007; pp. 927–941. [Google Scholar]
- Abbeloos, W. Real-Time Stereo Vision. Ph.D. Thesis, Karel de Grote-Hogeschool University College (KDG IWT), Antwerp, Belgium, 2010. [Google Scholar]
- Kumar, A.; Jain, P.K.; Pathak, P.M. Curve reconstruction of digitized surface using K-means algorithm. In Proceedings of the 24th DAAAM International Symposium on Intelligent Manufacturing and Automation, Univ Zadar, Zadar, Croatia, 23–26 October 2013; pp. 544–549. [Google Scholar]
- Bezdec, J.C. Pattern Recognition with Fuzzy Objective Function Algorithms; Plenum Press: New York, NY, USA, 1981. [Google Scholar]
- Teimouri, N.; Omid, M.; Mollazade, K.; Rajabipour, A. A novel artificial neural networks assisted segmentation algorithm for discriminating almond nut and shell from background and shadow. Comput. Electron. Agric. 2014, 105, 34–43. [Google Scholar] [CrossRef]


| Image | SAD | SSD | SGM | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Matching Window Size (Pixel) | Maximum Disparity (Pixel) | Matching Time (ms) | Matching Window Size (Pixel) | Maximum Disparity (Pixel) | Matching Time (ms) | Matching Window Size (Pixel) | Maximum Disparity (pixel) | Matching Time (ms) | |
| a | 55×55 | 130 | 1598 | 55×55 | 130 | 1610 | 15×15 | 128 | 142 |
| b | 55×55 | 120 | 1472 | 55×55 | 120 | 1502 | 15×15 | 128 | 135 |
| c | 55×55 | 110 | 1356 | 55×55 | 110 | 1383 | 15×15 | 128 | 138 |
| Image | Point Number of Original Point Cloud | Point Number of Point Cloud after Invalid Points Removal | Point Number of Sparse Point Cloud |
|---|---|---|---|
| a | 432,677 | 296,053 | 4096 |
| b | 432,677 | 277,858 | 4096 |
| c | 432,677 | 340,974 | 4096 |
| Image | GMM (ms) | K-means (ms) | Fuzzy c-means (ms) |
|---|---|---|---|
| a | 51 | 6 | 171 |
| b | 52 | 7 | 176 |
| c | 49 | 11 | 172 |
| Image | Area of Broccoli Seeding Obtained Manually (Pixel) | Area of Broccoli Seeding Obtained Theoretically (Pixel) | Intersection Area of Broccoli Seeding Obtained Manually and Theoretically (Pixel) | Sensitivity |
|---|---|---|---|---|
| a | 1.08 × 105 | 9.76 × 104 | 8.48 × 104 | 86.91% |
| b | 7.00 × 104 | 6.48 × 104 | 5.74 × 104 | 82.07% |
| c | 3.79 × 104 | 3.73 × 104 | 3.36 × 104 | 88.75% |
| Plants | Measured | Theoretical | (Theoretical Volume)/(Measured Volume) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Length (mm) | Width (mm) | Height (mm) | Volume (mm3) | Length (mm) | Width (mm) | Height (mm) | Volume (mm3) | ||
| a | 263.68 | 240.60 | 151.48 | 9.61 × 106 | 264.16 | 234.12 | 153.98 | 9.52 × 106 | 99.09% |
| b | 307.86 | 306.12 | 230.46 | 2.17 × 107 | 318.46 | 305.63 | 186.58 | 1.82 × 107 | 83.61% |
| c | 316.68 | 245.24 | 155.22 | 1.21 × 107 | 319.66 | 252.43 | 155.78 | 1.26 × 107 | 104.28% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, L.; Yang, Z.; Sun, Z.; Zhang, G.; Zhang, M.; Zhang, K.; Zhang, C.; Tan, Y.; Li, W. A Method for Broccoli Seedling Recognition in Natural Environment Based on Binocular Stereo Vision and Gaussian Mixture Model. Sensors 2019, 19, 1132. https://doi.org/10.3390/s19051132
Ge L, Yang Z, Sun Z, Zhang G, Zhang M, Zhang K, Zhang C, Tan Y, Li W. A Method for Broccoli Seedling Recognition in Natural Environment Based on Binocular Stereo Vision and Gaussian Mixture Model. Sensors. 2019; 19(5):1132. https://doi.org/10.3390/s19051132
Chicago/Turabian StyleGe, Luzhen, Zhilun Yang, Zhe Sun, Gan Zhang, Ming Zhang, Kaifei Zhang, Chunlong Zhang, Yuzhi Tan, and Wei Li. 2019. "A Method for Broccoli Seedling Recognition in Natural Environment Based on Binocular Stereo Vision and Gaussian Mixture Model" Sensors 19, no. 5: 1132. https://doi.org/10.3390/s19051132
APA StyleGe, L., Yang, Z., Sun, Z., Zhang, G., Zhang, M., Zhang, K., Zhang, C., Tan, Y., & Li, W. (2019). A Method for Broccoli Seedling Recognition in Natural Environment Based on Binocular Stereo Vision and Gaussian Mixture Model. Sensors, 19(5), 1132. https://doi.org/10.3390/s19051132





