Real-Time Prediction of Joint Forces by Motion Capture and Machine Learning
Abstract
:1. Introduction
2. Experimental Procedure and Biomechanical Simulations
2.1. Participants
2.2. Experimental Protocol
2.3. Musculoskeletal Modeling
2.4. Data Pre-Processing
3. Prediction by Machine Learning
3.1. Regression
3.2. Artificial Neural Networks
3.3. Support Vector Regression
3.4. Assessment
4. Results
4.1. Calculated KCFs Based on Musculoskeletal Modeling
4.2. ML-Based Prediction with and without GRFs
4.3. Robustness of Fit
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dreinhöfer, K.E.; Reichel, H.; Käfer, W. Lower limb pain. Best Pract. Res. Clin. Rheumatol. 2007, 21, 135–152. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.; Ingles, M.; Salvador-Pascual, A.; Cominetti, M.; Gomez-Cabrera, M.; Viña, J. Sarcopenia, frailty and their prevention by exercise. Free. Radic. Biol. Med. 2019, 132, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.A. Biomechanics of normal and pathological gait: Implications for understanding human locomotor control. J. Mot. Behav. 1989, 21, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Quadri, P.; Tettamanti, M.; Bernasconi, S.; Trento, F.; Loew, F. Lower limb function as predictor of falls and loss of mobility with social repercussions one year after discharge among elderly inpatients. Aging Clin. Exp. Res. 2005, 17, 82–89. [Google Scholar] [CrossRef]
- Neville, C.; Nguyen, H.; Ross, K.; Wingood, M.; Peterson, E.W.; DeWitt, J.E.; Moore, J.; King, M.J.; Atanelov, L.; White, J.; et al. Lower-Limb Factors Associated with Balance and Falls in Older Adults: A Systematic Review and Clinical Synthesis. J. Am. Podiatr. Med. Assoc. 2019, 110. [Google Scholar] [CrossRef]
- Steinman, B.A.; Pynoos, J.; Nguyen, A.Q.D. Fall risk in older adults: Roles of self-rated vision, home modifications, and limb function. J. Aging Health 2009, 21, 655–676. [Google Scholar] [CrossRef]
- Gregg, E.W.; Pereira, M.A.; Caspersen, C.J. Physical Activity, Falls, and Fractures Among Older Adults: A Review of the Epidemiologic Evidence. J. Am. Geriatr. Soc. 2000, 48, 883–893. [Google Scholar] [CrossRef]
- Hunter, D.J.; Schofield, D.J.; Callander, E.J. The individual and socioeconomic impact of osteoarthritis. Nat. Rev. Rheumatol. 2014, 10, 437–441. [Google Scholar] [CrossRef]
- Pisani, P.; Renna, M.D.; Conversano, F.; Casciaro, E.; Di Paola, M.; Quarta, E.; Muratore, M.; Casciaro, S. Major osteoporotic fragility fractures: Risk factor updates and societal impact. World J. Orthop. 2016, 7, 171–181. [Google Scholar] [CrossRef]
- Pedersen, B.K. The Physiology of Optimizing Health with a Focus on Exercise as Medicine. Annu. Rev. Physiol. 2019, 81, 607–627. [Google Scholar] [CrossRef]
- Lavie, C.J.; Arena, R.; Swift, D.L.; Johannsen, N.M.; Sui, X.; Lee, D.C.; Earnest, C.P.; Church, T.S.; O’Keefe, J.H.; Milani, R.V.; et al. Exercise and the cardiovascular system: Clinical science and cardiovascular outcomes. Circ. Res. 2015, 117, 207–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, B.G.; McInman, A. Exercise and the quality of life. Handb. Res. Sport Psychol. 1993, 729–760. [Google Scholar]
- Sañudo, B.; de Hoyo, M.; del Pozo-Cruz, J.; Carrasco, L.; del Pozo-Cruz, B.; Tejero, S.; Firth, E. A systematic review of the exercise effect on bone health: The importance of assessing mechanical loading in perimenopausal and postmenopausal women. Menopause 2017, 24, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Kersh, M.E.; Martelli, S.; Zebaze, R.; Seeman, E.; Pandy, M.G. Mechanical loading of the femoral neck in human locomotion. J. Bone Minber Res. 2018, 33, 1999–2006. [Google Scholar] [CrossRef] [Green Version]
- Carina, V.; Della Bella, E.; Costa, V.; Bellavia, D.; Veronesi, F.; Cepollaro, S.; Fini, M.; Giavaresi, G. Bone’s Response to Mechanical Loading in Aging and Osteoporosis: Molecular Mechanisms. Calcif. Tissue Int. 2020, 107, 301–318. [Google Scholar] [CrossRef]
- Musumeci, G. The Effect of Mechanical Loading on Articular Cartilage. J. Funct. Morphol. Kinesiol. 2016, 1, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Kinney, A.L.; Besier, T.F.; Silder, A.; Delp, S.L.; D’Lima, D.D.; Fregly, B.J. Changes in in vivo knee contact forces through gait modification. J. Orthop. Res. 2013, 31, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Simic, M.; Hinman, R.S.; Wrigley, T.V.; Bennell, K.L.; Hunt, M.A. Gait modification strategies for altering medial knee joint load: A systematic review. Arthritis Rheum. 2010, 63, 405–426. [Google Scholar] [CrossRef]
- Trepczynski, A.; Kutzner, I.; Kornaropoulos, E.; Taylor, W.R.; Duda, G.N.; Bergmann, G.; Heller, M.O. Patellofemoral joint contact forces during activities with high knee flexion. J. Orthop. Res. 2011, 30, 408–415. [Google Scholar] [CrossRef]
- Giarmatzis, G.; Jonkers, I.; Baggen, R.R.; Verschueren, S. Less hip joint loading only during running rather than walking in elderly compared to young adults. Gait Posture 2017, 53, 155–161. [Google Scholar] [CrossRef]
- Clouthier, A.L.; Smith, C.R.; Vignos, M.F.; Thelen, D.G.; Deluzio, K.J.; Rainbow, M.J. The effect of articular geometry features identified using statistical shape modelling on knee biomechanics. Med. Eng. Phys. 2019, 66, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, G.; De Groote, F.; Demeulenaere, B.; Mulier, M.; Van Der Perre, G.; Spaepen, A.; Jonkers, I. Subject-specific hip geometry affects predicted hip joint contact forces during gait. J. Biomech. 2008, 41, 1243–1252. [Google Scholar] [CrossRef]
- Halonen, K.S.; Dzialo, C.M.; Mannisi, M.; Venäläinen, M.S.; De Zee, M.; Andersen, M.S. Workflow assessing the effect of gait alterations on stresses in the medial tibial cartilage-combined musculoskeletal modelling and finite element analysis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Adouni, M.; Shirazi-Adl, A.; Shirazi, R. Computational biodynamics of human knee joint in gait: From muscle forces to cartilage stresses. J. Biomech. 2012, 45, 2149–2156. [Google Scholar] [CrossRef]
- Arokoski, J.P.A.; Jurvelin, J.S.; Väätäinen, U.; Helminen, H.J. Normal and pathological adaptations of articular cartilage to joint loading. Scand. J. Med. Sci. Sports 2000, 10, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Andriacchi, T.P.; Mündermann, A. The role of ambulatory mechanics in the initiation and progression of knee osteoarthritis. Curr. Opin. Rheumatol. 2006, 18, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Herzog, W.; Epstein, M. Joint contact mechanics in the early stages of osteoarthritis. Med. Eng. Phys. 2000, 22, 1–12. [Google Scholar] [CrossRef]
- D’Lima, D.D.; Fregly, B.J.; Patil, S.; Steklov, N.; Colwell, J.C.W. Knee joint forces: Prediction, measurement, and significance. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2012, 226, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Fregly, B.J. Gait Modification to Treat Knee Osteoarthritis. HSS J. 2012, 8, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Zingde, S.M.; Slamin, J. Biomechanics of the knee joint, as they relate to arthroplasty. Orthop. Trauma 2017, 31, 1–7. [Google Scholar] [CrossRef]
- Andriacchi, T.P.; Hurwitz, D.E. Gait biomechanics and the evolution of total joint replacement. Gait Posture 1997, 5, 256–264. [Google Scholar] [CrossRef]
- Felson, D.T.; Nevitt, M.C.; Zhang, Y.; Aliabadi, P.; Baumer, B.; Gale, D.; Li, W.; Yu, W.; Xu, L. High prevalence of lateral knee osteoarthritis in Beijing Chinese compared with Framingham Caucasian subjects. Arthritis Rheum. 2002, 46, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Eddo, O.; Lindsey, B.; Caswell, S.V.; Cortes, N. Current Evidence of Gait Modification with Real-time Biofeedback to Alter Kinetic, Temporospatial, and Function-Related Outcomes: A Review. Int. J. Kinesiol. Sports Sci. 2017, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Noort, J.C.V.D.; Steenbrink, F.; Roeles, S.; Harlaar, J. Real-time visual feedback for gait retraining: Toward application in knee osteoarthritis. Med. Biol. Eng. Comput. 2015, 53, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Hilding, M.B.; Lanshammar, H.; Ryd, L. Knee joint loading and tibial component loosening. RSA and gait analysis in 45 osteoarthritic patients before and after TKA. J. Bone Jt. Surg. Ser. B 1996, 78, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Komistek, R.D.; Kane, T.R.; Mahfouz, M.; Ochoa, J.A.; Dennis, D.A. Knee mechanics: A review of past and present techniques to determine in vivo loads. J. Biomech. 2005, 38, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Al Nazer, R.; Lanovaz, J.; Kawalilak, C.; Johnston, J.; Kontulainen, S. Direct in vivo strain measurements in human bone—A systematic literature review. J. Biomech. 2012, 45, 27–40. [Google Scholar] [CrossRef]
- Papagiannaki, A.; Zacharaki, E.I.; Kalouris, G.; Kalogiannis, S.; Deltouzos, K.; Ellul, J.; Megalooikonomou, V. Recognizing Physical Activity of Older People from Wearable Sensors and Inconsistent Data. Sensors 2019, 19, 880. [Google Scholar] [CrossRef] [Green Version]
- Pellicer-Valero, O.J.; Rupérez, M.J.; Martínez-Sanchis, S.; Martín-Guerrero, J.D. Real-time biomechanical modeling of the liver using Machine Learning models trained on Finite Element Method simulations. Expert Syst. Appl. 2020, 143, 113083. [Google Scholar] [CrossRef]
- Mundt, M.; Thomsen, W.; Witter, T.; Koeppe, A.; David, S.; Bamer, F.; Potthast, W.; Markert, B. Prediction of lower limb joint angles and moments during gait using artificial neural networks. Med. Biol. Eng. Comput. 2020, 58, 211–225. [Google Scholar] [CrossRef]
- Ardestani, M.M.; Zhang, X.; Wang, L.; Lian, Q.; Liu, Y.; He, J.; Li, D.; Jin, Z. Human lower extremity joint moment prediction: A wavelet neural network approach. Expert Syst. Appl. 2014, 41, 4422–4433. [Google Scholar] [CrossRef]
- Favre, J.; Hayoz, M.; Erhart-Hledik, J.C.; Andriacchi, T.P. A neural network model to predict knee adduction moment during walking based on ground reaction force and anthropometric measurements. J. Biomech. 2012, 45, 692–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, L.; Jones, R.K.; Howard, D. Whole body inverse dynamics over a complete gait cycle based only on measured kinematics. J. Biomech. 2008, 41, 2750–2759. [Google Scholar] [CrossRef]
- Lim, H.; Kim, B.; Park, S. Prediction of Lower Limb Kinetics and Kinematics during Walking by a Single IMU on the Lower Back Using Machine Learning. Sensors 2020, 20, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, T.; Kwon, H.; Oh, S.E.; Joo, S.-B.; Choi, A.; Heo, H.M.; Kim, K.; Mun, J.H. Predicting Complete Ground Reaction Forces and Moments During Gait With Insole Plantar Pressure Information Using a Wavelet Neural Network. J. Biomech. Eng. 2015, 137, 091001. [Google Scholar] [CrossRef]
- De Brabandere, A.; Emmerzaal, J.; Timmermans, A.; Jonkers, I.; Vanwanseele, B.; Davis, J. A Machine Learning Approach to Estimate Hip and Knee Joint Loading Using a Mobile Phone-Embedded IMU. Front. Bioeng. Biotechnol. 2020, 8, 320. [Google Scholar] [CrossRef] [Green Version]
- Rane, L.; Ding, Z.; McGregor, A.H.; Bull, A.M.J. Deep Learning for Musculoskeletal Force Prediction. Ann. Biomed. Eng. 2019, 47, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Ardestani, M.M.; Chen, Z.; Wang, L.; Lian, Q.; Liu, Y.; He, J.; Li, D.; Jin, Z. Feed forward artificial neural network to predict contact force at medial knee joint: Application to gait modification. Neurocomputing 2014, 139, 114–129. [Google Scholar] [CrossRef]
- Cleather, D.J. Neural network based approximation of muscle and joint contact forces during jumping and landing. J. Hum. Perform. Health 2019, 1, 1–13. [Google Scholar]
- Ardestani, M.M.; Moazen, M.; Chen, Z.; Zhang, J.; Jin, Z. A real-time topography of maximum contact pressure distribution at medial tibiofemoral knee implant during gait: Application to knee rehabilitation. Neurocomputing 2015, 154, 174–188. [Google Scholar] [CrossRef]
- Lu, Y.; Pulasani, P.R.; Derakhshani, R.; Guess, T.M. Application of neural networks for the prediction of cartilage stress in a musculoskeletal system. Biomed. Signal Process. Control 2013, 8, 475–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hambli, R. Application of Neural Networks and Finite Element Computation for Multiscale Simulation of Bone Remodeling. J. Biomech. Eng. 2010, 132, 114502. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Chu, C.C.; Chow, D.H. EMG-based lumbosacral joint compression force prediction using a support vector machine. Med. Eng. Phys. 2019, 74, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Yoshida, T.; Ernst, M.; Schmalz, T.; Schilling, A.F. A support vector regression approach for continuous prediction of joint angles and moments during walking: An implication for controlling active knee-ankle prostheses/orthoses. In Proceedings of the 2019 IEEE International Conference on Cyborg and Bionic Systems (CBS), Munich, Germany, 18–20 September 2019; pp. 66–71. [Google Scholar]
- Ahuja, S.; Jirattigalachote, W.; Tosborvorn, A. Improving Accuracy of Inertial Measurement Units using Support Vector Regression. Cs229.Stanford.Edu. 2011. Available online: http://www.wisitmax.com/img/researchexp/stanford/wearablegait/cs229poster.pdf (accessed on 3 December 2020).
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef]
- Lerner, Z.F.; Demers, M.S.; Delp, S.L.; Browning, R.C. How tibiofemoral alignment and contact locations affect predictions of medial and lateral tibiofemoral contact forces. J. Biomech. 2015, 48, 644–650. [Google Scholar] [CrossRef] [Green Version]
- Haykin, S. Neural Networks and Learning Machines, 3rd ed.; Pearson Education India: Delhi, India, 2010; Available online: http://repository.fue.edu.eg/xmlui/bitstream/handle/123456789/3421/5618.pdf?sequence=1 (accessed on 3 December 2020).
- Drucker, H.; Burges, C.J.C.; Kaufman, L.; Smola, A.J.; Vapnik, V. Support vector regression machines. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 1997; pp. 155–161. [Google Scholar]
- Preece, S.J.; Goulermas, J.Y.; Kenney, L.P.J.; Howard, D.; Meijer, K.; Crompton, R. Activity identification using body-mounted sensors—A review of classification techniques. Physiol. Meas. 2009, 30, R1–R33. [Google Scholar] [CrossRef]
- Evrigkas, M.; Nikou, C.; Kakadiaris, I.A. A Review of Human Activity Recognition Methods. Front. Robot. AI 2015, 2, 28. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- Stetter, B.J.; Ringhof, S.; Krafft, F.C.; Sell, S.; Stein, T. Estimation of Knee Joint Forces in Sport Movements Using Wearable Sensors and Machine Learning. Sensors 2019, 19, 3690. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Xu, W.; Luo, G.; Wang, H.; Yang, J.; Lu, W. Random Forest enhancement using improved Artificial Fish Swarm for the medial knee contact force prediction. Artif. Intell. Med. 2020, 103, 101811. [Google Scholar] [CrossRef]
- Kristianslund, E.; Krosshaug, T.; Bogert, A.J.V.D. Effect of low pass filtering on joint moments from inverse dynamics: Implications for injury prevention. J. Biomech. 2012, 45, 666–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattos, C.L.C.; Dai, Z.; Damianou, A.; Barreto, G.A.; Lawrence, N.D. Deep recurrent Gaussian processes for outlier-robust system identification. J. Process. Control 2017, 60, 82–94. [Google Scholar] [CrossRef]
- Horst, F.; Lapuschkin, S.; Samek, W.; Müller, K.-R.; Schöllhorn, W.I. Explaining the unique nature of individual gait patterns with deep learning. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L.; Friedman, J.H. Predicting Multivariate Responses in Multiple Linear Regression. J. R. Stat. Soc. Ser. B Stat. Methodol. 1997, 59, 3–54. [Google Scholar] [CrossRef]
- Fregly, B.J.; Besier, T.F.; Lloyd, D.G.; Delp, S.L.; Banks, S.A.; Pandy, M.G.; D’Lima, D.D. Grand challenge competition to predict in vivo knee loads. J. Orthop. Res. 2012, 30, 503–513. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Banks, S.A.; Mitchell, K.H.; D’lima, D.D.; Colwell, C.W.; Fregly, B.J. Correlation between the knee adduction torque and medial contact force for a variety of gait patterns. J. Orthop. Res. 2007, 25, 789–797. [Google Scholar] [CrossRef]
- Kutzner, I.; Küther, S.; Heinlein, B.; Dymke, J.; Bender, A.; Halder, A.M.; Bergmann, G. The effect of valgus braces on medial compartment load of the knee joint–in vivo load measurements in three subjects. J. Biomech. 2011, 44, 1354–1360. [Google Scholar] [CrossRef]
- Winby, C.; Lloyd, D.; Besier, T.; Kirk, T. Muscle and external load contribution to knee joint contact loads during normal gait. J. Biomech. 2009, 42, 2294–2300. [Google Scholar] [CrossRef]
- Kumar, D.; Manal, K.; Rudolph, K.S. Knee joint loading during gait in healthy controls and individuals with knee osteoarthritis. Osteoarthr. Cartil. 2013, 21, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Giarmatzis, G.; Jonkers, I.; Wesseling, M.; Van Rossom, S.; Verschueren, S. Loading of Hip Measured by Hip Contact Forces at Different Speeds of Walking and Running. J. Bone Miner. Res. 2015, 30, 1431–1440. [Google Scholar] [CrossRef]
- Matijevich, E.S.; Branscombe, L.M.; Scott, L.R.; Zelik, K.E. Ground reaction force metrics are not strongly correlated with tibial bone load when running across speeds and slopes: Implications for science, sport and wearable tech. PLoS ONE 2019, 14, e0210000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolopoulos, F.P.; Zacharaki, E.I.; Moustakas, K.; Moustakas, K. Personalized Knee Geometry Modeling Based on Multi-Atlas Segmentation and Mesh Refinement. IEEE Access 2020, 8, 56766–56781. [Google Scholar] [CrossRef]
- Stanev, D.; Moustakas, K. Modeling musculoskeletal kinematic and dynamic redundancy using null space projection. PLoS ONE 2019, 14, e0209171. [Google Scholar]
- Ntalianis, V.; Fakotakis, N.D.; Nousias, S.; Lalos, A.S.; Birbas, M.; Zacharaki, E.; Moustakas, K. Deep CNN Sparse Coding for Real Time Inhaler Sounds Classification. Sensors 2020, 20, 2363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Zhang, Y.; Cheng, J.; Lu, H. Skeleton-based action recognition with directed graph neural networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 7904–7913. [Google Scholar]





| No. Subjects | Mean Age (SD) (Years) | Body Mass Index (kg/m2) | Percent Female | ||||
|---|---|---|---|---|---|---|---|
| Young 40 | Elderly 14 | Young 22 (1.66) | Elderly 69.6 (3.5) | Young 22.8 (2.8) | Elderly 24.4 (2.3) | Young 60% | Elderly 100% |
| Total = 54 | |||||||
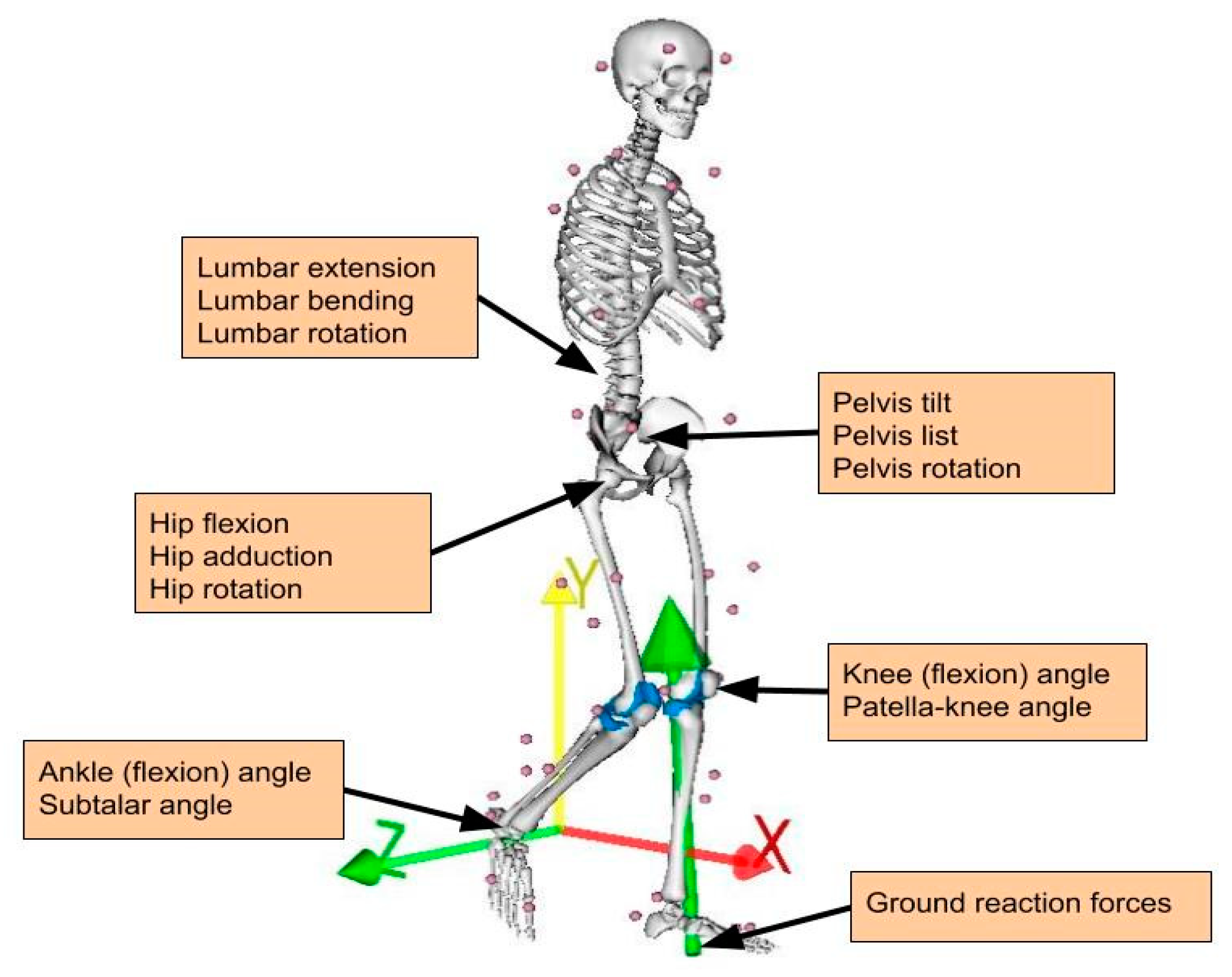
| Anatomical Location | Abbreviation | Component | Units |
|---|---|---|---|
| torso (lumbrosacral joint) | lumbar | extension bending rotation | degrees |
| pelvis | pelvis | tilt list rotation | |
| hip joint | hip | flexion adduction rotation | |
| knee joint | knee | flexion | |
| patella knee angle | patella | flexion | |
| ankle joint | ankle | flexion | |
| subtalar joint | subtalar | eversion | |
| Force Description | |||
| ground reaction force | GRF | anteroposterior (x) distal proximal (y) mediolateral (z) | body weight (BW) |
| medial knee contact force | KCF (med) | anteroposterior (x) distal proximal (y) mediolateral (z) | |
| lateral knee contact force | KCF (lat) | anteroposterior (x) distal proximal (y) mediolateral (z) |
| Medial Force | Lateral Force | |||
|---|---|---|---|---|
| 1st Peak | 2nd Peak | 1st Peak | 2nd Peak | |
| 3 km/h | 2.18 (0.58) | 2.50 (0.75) | 0.71(0.34) | 0.70 (0.39) |
| 4 km/h | 2.22 (0.58) | 2.76 (0.82) | 0.92 (0.45) | 0.81 (0.49) |
| 5 km/h | 2.48 (0.63) | 3.06 (0.86) | 1.15 (0.49) | 1.02 (0.49) |
| 6 km/h | 2.82 (0.71) | 3.25 (0.96) | 1.44 (0.63) | 1.36 (0.79) |
| 7 km/h | 3.30 (0.65) | 3.22 (1.05) | 1.84 (0.68) | 1.73 (0.77) |
| med_x | med_y | med_z | lat_y | lat_z | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LeaveTrialsOut | ||||||||||
| GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | |
| SVR | 1.04 | 1.36 | 2.68 | 3.92 | 2.03 | 2.65 | 2.80 | 3.44 | 2.79 | 3.44 |
| ANN | 0.67 | 0.9 | 1.71 | 2.35 | 1.45 | 1.82 | 1.61 | 2.04 | 1.61 | 2.04 |
| LeaveSubjectsOut | ||||||||||
| GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | |
| SVR | 1.53 | 1.73 | 4.63 | 5.85 | 3.42 | 3.80 | 4.41 | 4.66 | 4.41 | 4.65 |
| ANN | 1.60 | 1.81 | 4.54 | 5.39 | 3.49 | 3.85 | 4.19 | 4.59 | 4.19 | 4.59 |
| med_x | med_y | med_z | lat_y | lat_z | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LeaveTrialsOut | ||||||||||
| GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | |
| SVR | 0.85 | 0.73 | 0.94 | 0.88 | 0.94 | 0.89 | 0.83 | 0.73 | 0.83 | 0.73 |
| ANN | 0.94 | 0.89 | 0.98 | 0.96 | 0.97 | 0.95 | 0.95 | 0.91 | 0.95 | 0.91 |
| LeaveSubjectsOut | ||||||||||
| GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | GRF | noGRF | |
| SVR | 0.64 | 0.50 | 0.83 | 0.73 | 0.82 | 0.77 | 0.52 | 0.44 | 0.52 | 0.44 |
| ANN | 0.63 | 0.48 | 0.85 | 0.76 | 0.83 | 0.76 | 0.58 | 0.45 | 0.58 | 0.45 |
| Subjects | Test Trials | Classifier | Inputs | Y 2 | Mean Pearson’s R (NRMSE%) 3 | ||
|---|---|---|---|---|---|---|---|
| LeaveTrialsOut | LeaveSubjectsOut | ||||||
| Ardestani et al. (2014) [48] | 4 (knee replacement patients) | 75 | ANN | -GRFs -marker 3D coordinates -EMG | in vivo | 0.96 (10.5) | 0.94 (13.3) |
| Rane et al. (2019) [47] | healthy and knee OA patients | 58 63 28 | CNN | -CoP 1 -GRFs -marker 3D coordinates | ID | 0.90 | 0.90 0.87 |
| Stetter et al. (2019) [65] | 13 healthy athletes (young) | 198 | ANN | -2 IMUs | ID | - | 0.87 |
| Zhu et al. (2019) [66] | 3 (knee replacement patients) | 135 | Random Forest | -GRFs -marker 3D coordinates -EMG | in vivo | 0.97 | - |
| Proposed method | 54 healthy (young and elderly) | 4784 | ANN | -GRFs -Joint angles | ID | 0.98 (1.71) | 0.85 (4.54) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giarmatzis, G.; Zacharaki, E.I.; Moustakas, K. Real-Time Prediction of Joint Forces by Motion Capture and Machine Learning. Sensors 2020, 20, 6933. https://doi.org/10.3390/s20236933
Giarmatzis G, Zacharaki EI, Moustakas K. Real-Time Prediction of Joint Forces by Motion Capture and Machine Learning. Sensors. 2020; 20(23):6933. https://doi.org/10.3390/s20236933
Chicago/Turabian StyleGiarmatzis, Georgios, Evangelia I. Zacharaki, and Konstantinos Moustakas. 2020. "Real-Time Prediction of Joint Forces by Motion Capture and Machine Learning" Sensors 20, no. 23: 6933. https://doi.org/10.3390/s20236933





