LeafScope: A Portable High-Resolution Multispectral Imager for In Vivo Imaging Soybean Leaf
Abstract
1. Introduction
2. Development of LeafScope
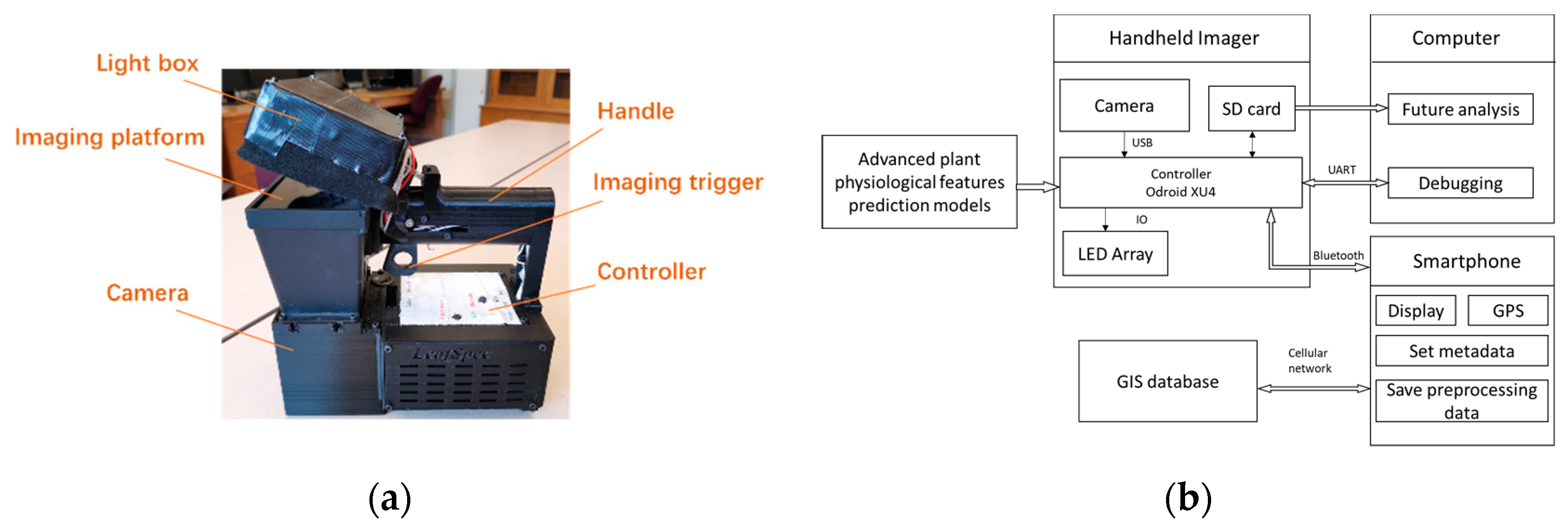
2.1. Hardware Configuration
2.2. Workflow of LeafScope
2.3. Image Processing
2.3.1. Spectral Calibration
2.3.2. Image Segmentation
2.3.3. Color-based and Morphological Features Extraction
2.4. LeafSpec App
3. Test of LeafScope
3.1. Plant Samples
3.2. Data Acquisition
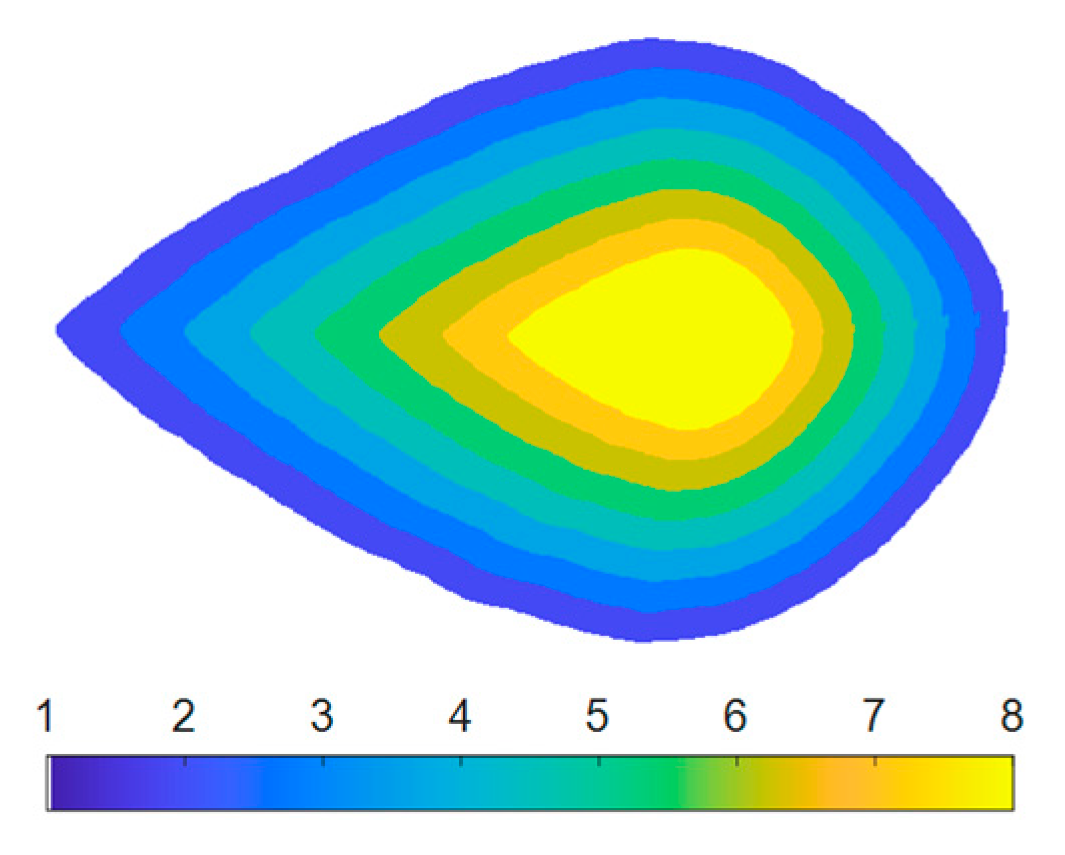
3.3. Color Distribution across the Leaf
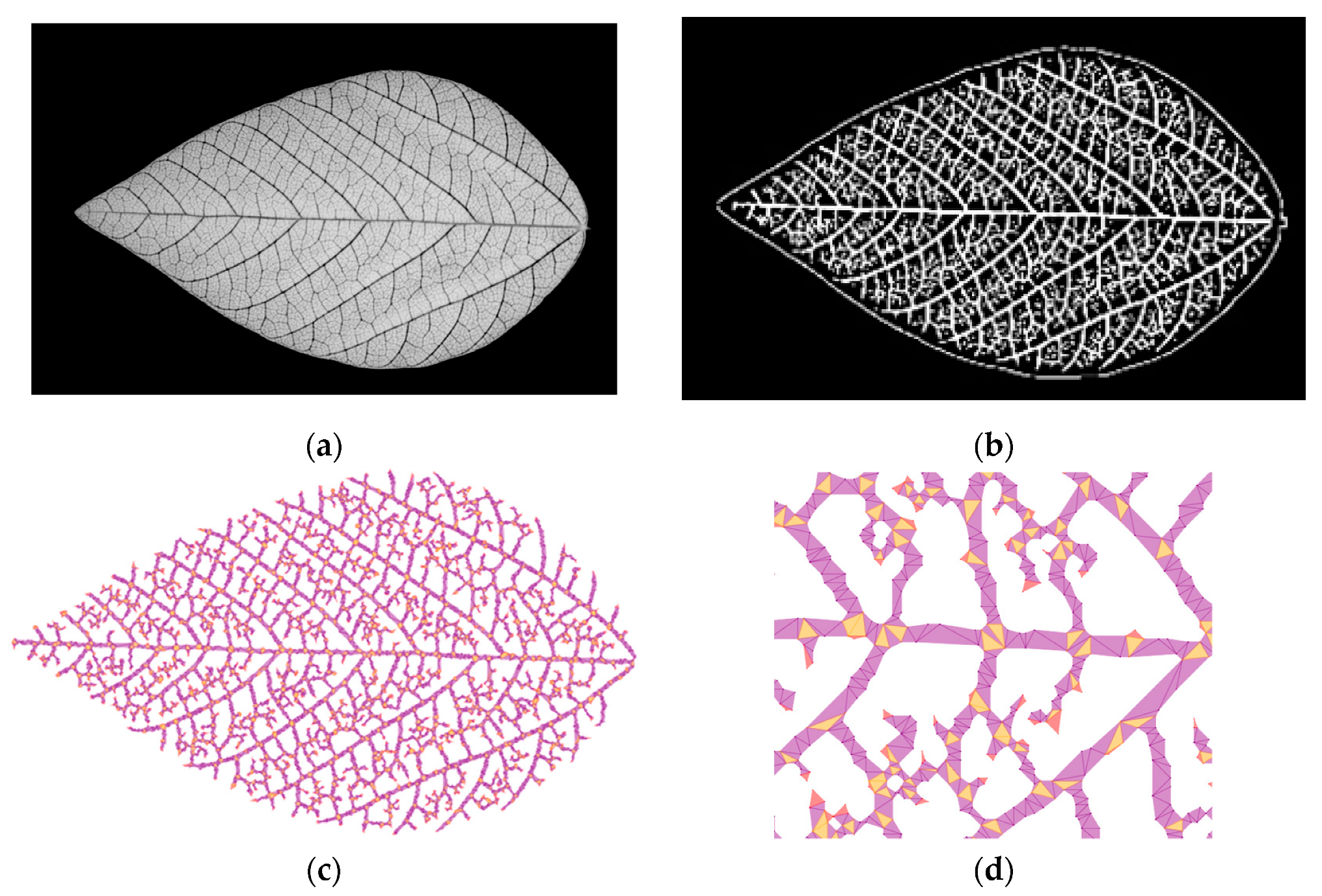
3.4. Leaf Venation Features Extraction
3.5. Statistical Analysis
4. Results and Discussions
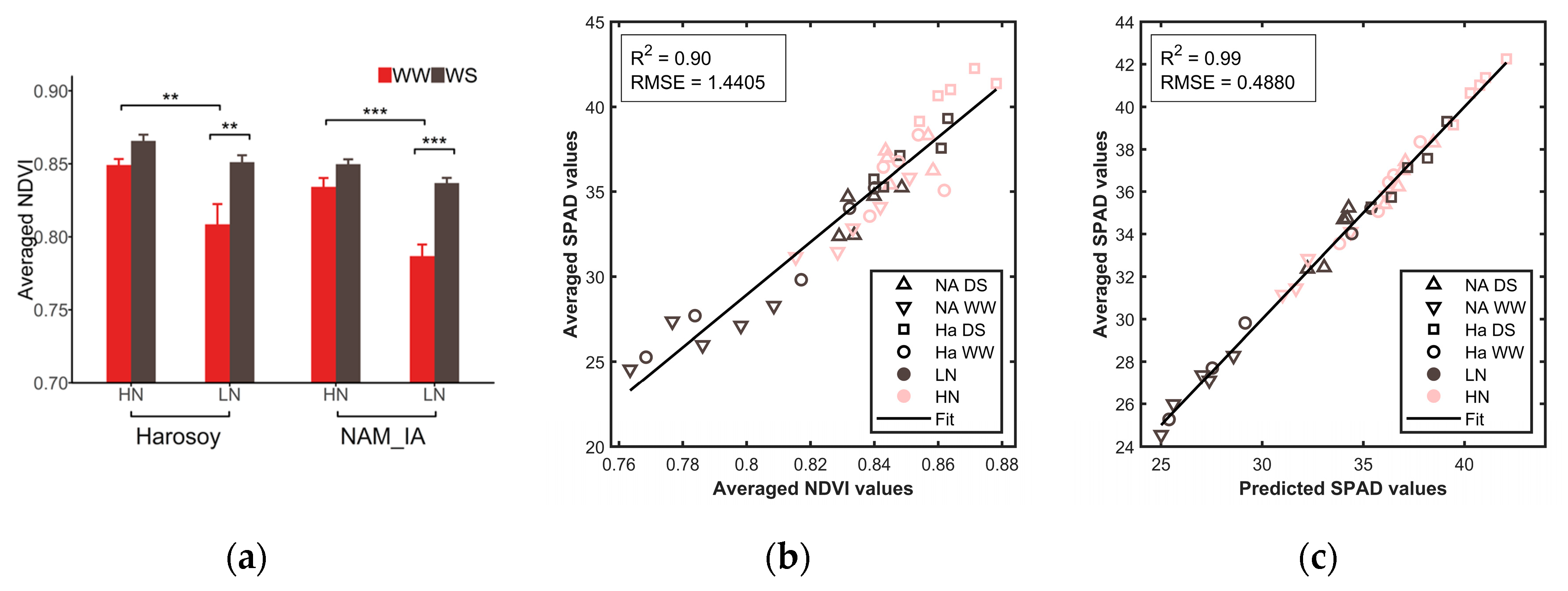
4.1. Color Features
4.1.1. Averaged Color Features of Whole Leaves
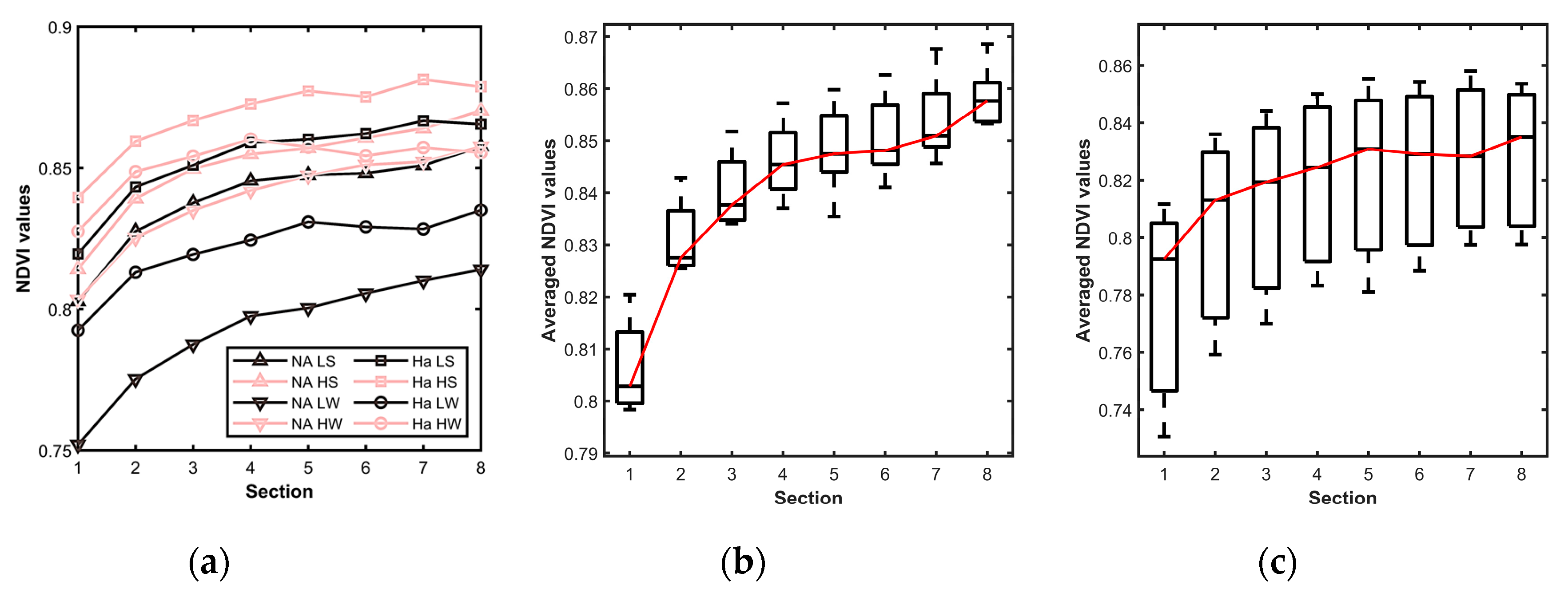
4.1.2. Color Distribution across the Leaf
4.2. Leaf Morphological Features
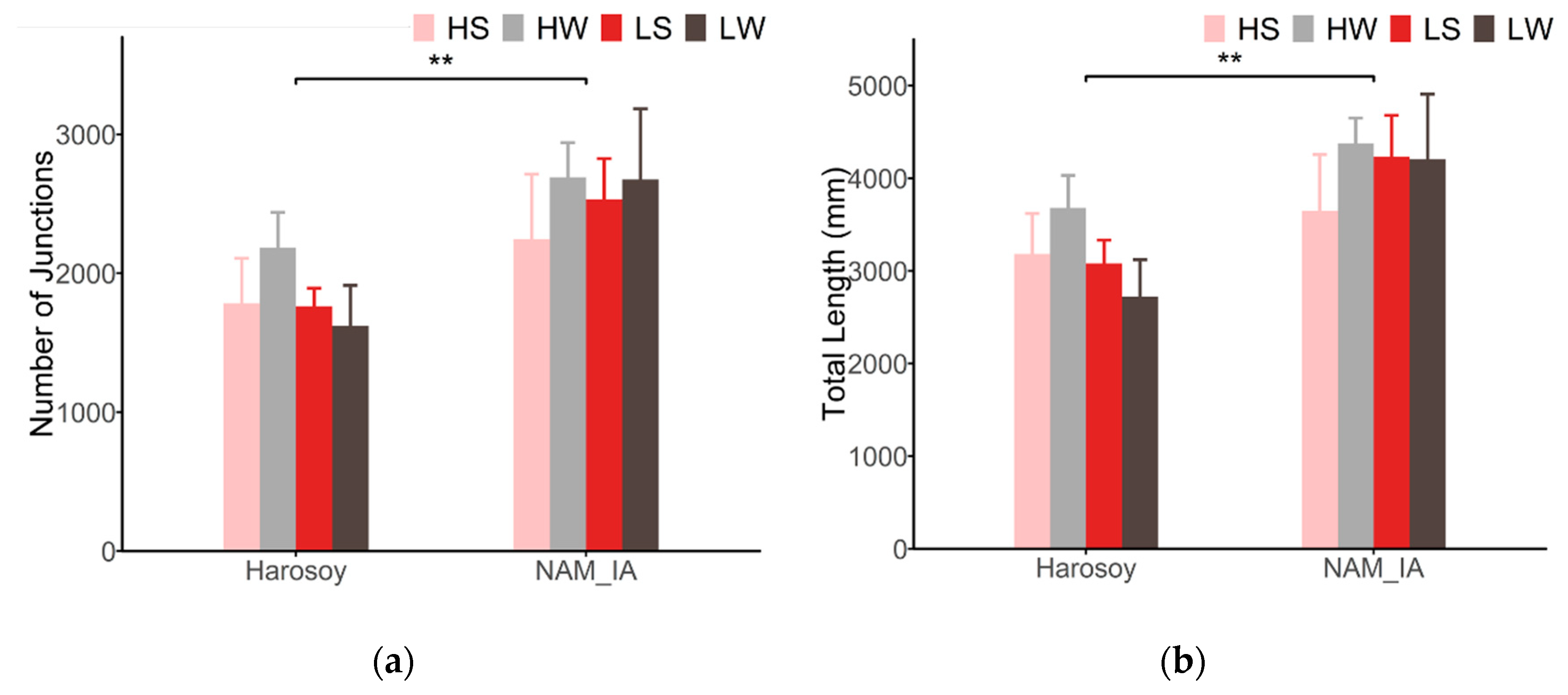
4.3. Leaf Venation Features
4.4. Opportunities for Future Work
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baerenfaller, K.; Massonnet, C.; Walsh, S.; Baginsky, S.; Bühlmann, P.; Hennig, L.; Hirsch-Hoffmann, M.; Howell, K.A.; Kahlau, S.; Radziejwoski, A.; et al. Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol. Syst. Biol. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 2013, 18, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sharma, M.K. Nutrient deficiencies of field crops: guide to diagnosis and management; Kumar, P., Sharma, M.K., Eds.; Cabi: Wallingford, UK, 2013; ISBN 9781780642789. [Google Scholar]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef] [PubMed]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef]
- Maharlooei, M.; Sivarajan, S.; Bajwa, S.G.; Harmon, J.P.; Nowatzki, J. Detection of soybean aphids in a greenhouse using an image processing technique. Comput. Electron. Agric. 2017, 132, 63–70. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Q.; Wu, B.; Zhu, Y.; Liu, D.; Zhang, J.; Jin, Z. A review of leaf morphology plasticity linked to plant response and adaptation characteristics in arid ecosystems. Chin. J. Plant Ecol. 2012, 36, 88–98. [Google Scholar] [CrossRef]
- Roth-Nebelsick, A.; Uhl, D.; Mosbrugger, V.; Kerp, H. Evolution and function of leaf venation architecture: A review. Ann. Bot. 2001, 87, 553–566. [Google Scholar] [CrossRef]
- Price, C.A.; Symonova, O.; Mileyko, Y.; Hilley, T.; Weitz, J.S. Leaf extraction and analysis framework graphical user interface: Segmenting and analyzing the structure of leaf veins and areoles. Plant Physiol. 2011, 155, 236–245. [Google Scholar] [CrossRef]
- Bühler, J.; Rishmawi, L.; Pflugfelder, D.; Huber, G.; Scharr, H.; Hülskamp, M.; Koornneef, M.; Schurr, U.; Jahnke, S. Phenovein—A tool for leaf vein segmentation and analysis. Plant Physiol. 2015, 169, 2359–2370. [Google Scholar] [CrossRef]
- Lasser, J.; Katifori, E. NET: a new framework for the vectorization and examination of network data. Source Code Biol. Med. 2017, 12, 4. [Google Scholar] [CrossRef]
- Dhondt, S.; Van Haerenborgh, D.; Van Cauwenbergh, C.; Merks, R.M.H.; Philips, W.; Beemster, G.T.S.; Inzé, D. Quantitative analysis of venation patterns of Arabidopsis leaves by supervised image analysis. Plant J. 2012, 69, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Dirnberger, M.; Kehl, T.; Neumann, A. NEFI: Network extraction from images. Sci. Rep. 2015, 5, 15669. [Google Scholar] [CrossRef]
- Gan, Y.; Rong, Y.; Huang, F.; Hu, L.; Yu, X.; Duan, P.; Xiong, S.; Liu, H.; Peng, J.; Yuan, X. Automatic hierarchy classification in venation networks using directional morphological filtering for hierarchical structure traits extraction. Comput. Biol. Chem. 2019, 80, 187–194. [Google Scholar] [CrossRef]
- Bradstreet, R.B. Kjeldahl Method for Organic Nitrogen. Anal. Chem. 1954, 26, 185–187. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Bloom, P.R. Extinction coefficients of chlorophyll a and b in N, N -dimethylformamide and 80% acetone. Plant Physiol. 1985, 77, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Dillenburg, L.R. Measurement of leaf relative water content in Araucaria Angustifolia. Rev. Bras. De Fisiol. Veg. 1999, 11, 69–75. [Google Scholar]
- Goetz, A.F.H. Three decades of hyperspectral remote sensing of the Earth: A personal view. Remote Sens. Environ. 2009, 113, S5–S16. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Wendel, A.; Underwood, J. Ground based hyperspectral imaging for extensive mango yield estimation. Comput. Electron. Agric. 2019, 157, 126–135. [Google Scholar] [CrossRef]
- Purdue University Controlled Environment Phenotyping Facility. Available online: https://ag.purdue.edu/cepf/ (accessed on 5 September 2019).
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef]
- Markwell, J.; Osterman, J.C.; Mitchell, J.L. Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynth. Res. 1995, 46, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Chen, J.; Yu, T.; Gao, W.; Ling, X.; Li, Y.; Peng, S.; Huang, J. SPAD-based leaf nitrogen estimation is impacted by environmental factors and crop leaf characteristics. Sci. Rep. 2015, 5, 13389. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Atefi, A.; Zhang, H.; Miao, C.; Ramamurthy, R.K.; Sigmon, B.; Yang, J.; Schnable, J.C. High-throughput analysis of leaf physiological and chemical traits with VIS–NIR–SWIR spectroscopy: a case study with a maize diversity panel. Plant Methods 2019, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Ghazal, M.; Mahmoud, A.; Shalaby, A.; El-Baz, A. Automated framework for accurate segmentation of leaf images for plant health assessment. Environ. Monit. Assess. 2019, 191, 491. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Detection of nutrition deficiencies in plants using proximal images and machine learning: A review. Comput. Electron. Agric. 2019, 162, 482–492. [Google Scholar] [CrossRef]
- Wang, L.; Jin, J.; Song, Z.; Wang, J.; Zhang, L.; Rehman, T.U.; Ma, D.; Carpenter, N.R.; Tuinstra, M.R. LeafSpec: An accurate and portable hyperspectral corn leaf imager. Comput. Electron. Agric. 2020, 169. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Wang, J.; Song, Z.; Rehman, T.U.; Bureetes, T.; Ma, D.; Chen, Z.; Neeno, S.; Jin, J. Leaf Scanner: A portable and low-cost multispectral corn leaf scanning device for precise phenotyping. Comput. Electron. Agric. 2019, 167, 105069. [Google Scholar] [CrossRef]
- Tirado, S.B.; Dennis, S.S.; Enders, T.A.; Springer, N.M. Utilizing top-down hyperspectral imaging for monitoring genotype and growth conditions in maize. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Manivannan, P.; Wahid, A.; Farooq, M.; Al-Juburi, H.J.; Somasundaram, R.; Panneerselvam, R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int. J. Agric. Biol. 2009, 11, 100–105. [Google Scholar]
- Schneider, J.V.; Rabenstein, R.; Wesenberg, J.; Wesche, K.; Zizka, G.; Habersetzer, J. Improved non-destructive 2D and 3D X-ray imaging of leaf venation. Plant Methods 2018, 14, 7. [Google Scholar] [CrossRef] [PubMed]






| Component | Price ($) | Parameters | Values |
|---|---|---|---|
| Camera | 519 | Model | BFLY-U3-23S6M-C |
| Resolution | 1920 × 1200 | ||
| Frame rate | 41 FPS | ||
| Chroma | Mono | ||
| Sensor mode | Sony IMX249 | ||
| Pixel size | 5.86 µm | ||
| Shutter type | Global | ||
| ADC | 12 bit | ||
| Dynamic range (dB) | 67.12 | ||
| Dimensions (mm) | 29 × 29 × 30 | ||
| Mass (g) | 36 | ||
| Lens | 379 | Model | V0814-MP |
| Focal length | 8 mm | ||
| LED 405 nm | 24 | FWHM | 40 nm |
| LED 560 nm | 10 | FWHM | 24 nm |
| LED 660 nm | 5 | FWHM | 38 nm |
| LED 860 nm | 14 | FWHM | 50 nm |
| Controller | 49 | Model | ODROID XU4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Duan, Y.; Zhang, L.; Wang, J.; Li, Y.; Jin, J. LeafScope: A Portable High-Resolution Multispectral Imager for In Vivo Imaging Soybean Leaf. Sensors 2020, 20, 2194. https://doi.org/10.3390/s20082194
Wang L, Duan Y, Zhang L, Wang J, Li Y, Jin J. LeafScope: A Portable High-Resolution Multispectral Imager for In Vivo Imaging Soybean Leaf. Sensors. 2020; 20(8):2194. https://doi.org/10.3390/s20082194
Chicago/Turabian StyleWang, Liangju, Yunhong Duan, Libo Zhang, Jialei Wang, Yikai Li, and Jian Jin. 2020. "LeafScope: A Portable High-Resolution Multispectral Imager for In Vivo Imaging Soybean Leaf" Sensors 20, no. 8: 2194. https://doi.org/10.3390/s20082194
APA StyleWang, L., Duan, Y., Zhang, L., Wang, J., Li, Y., & Jin, J. (2020). LeafScope: A Portable High-Resolution Multispectral Imager for In Vivo Imaging Soybean Leaf. Sensors, 20(8), 2194. https://doi.org/10.3390/s20082194






