Deep Learning Based Evaluation of Spermatozoid Motility for Artificial Insemination
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Image Annotation
2.3. Neural Network
- The number of classes is 1 because in the Tensorflow API (Python) the background is not considered as a class, and only objects themselves are counted as a single class;
- The maximum number of first stage proposals—since there can be an average of 30 sperm per frame (the value ranges from several units to 100), the selected value is 100;
- The maximum number of detections per class—the default value is 100. Since there can be on average 30 sperm per frame (value ranges from several units to 100), the selected value is 100;
- The maximum number of total detections—the default value is 100. The project includes one class, depending on the maximum number of detections per class, the selected value is 100;
- Score converter—the sigmoid function is used.
2.4. Spermatozoid Tracking Algorithm
3. Results
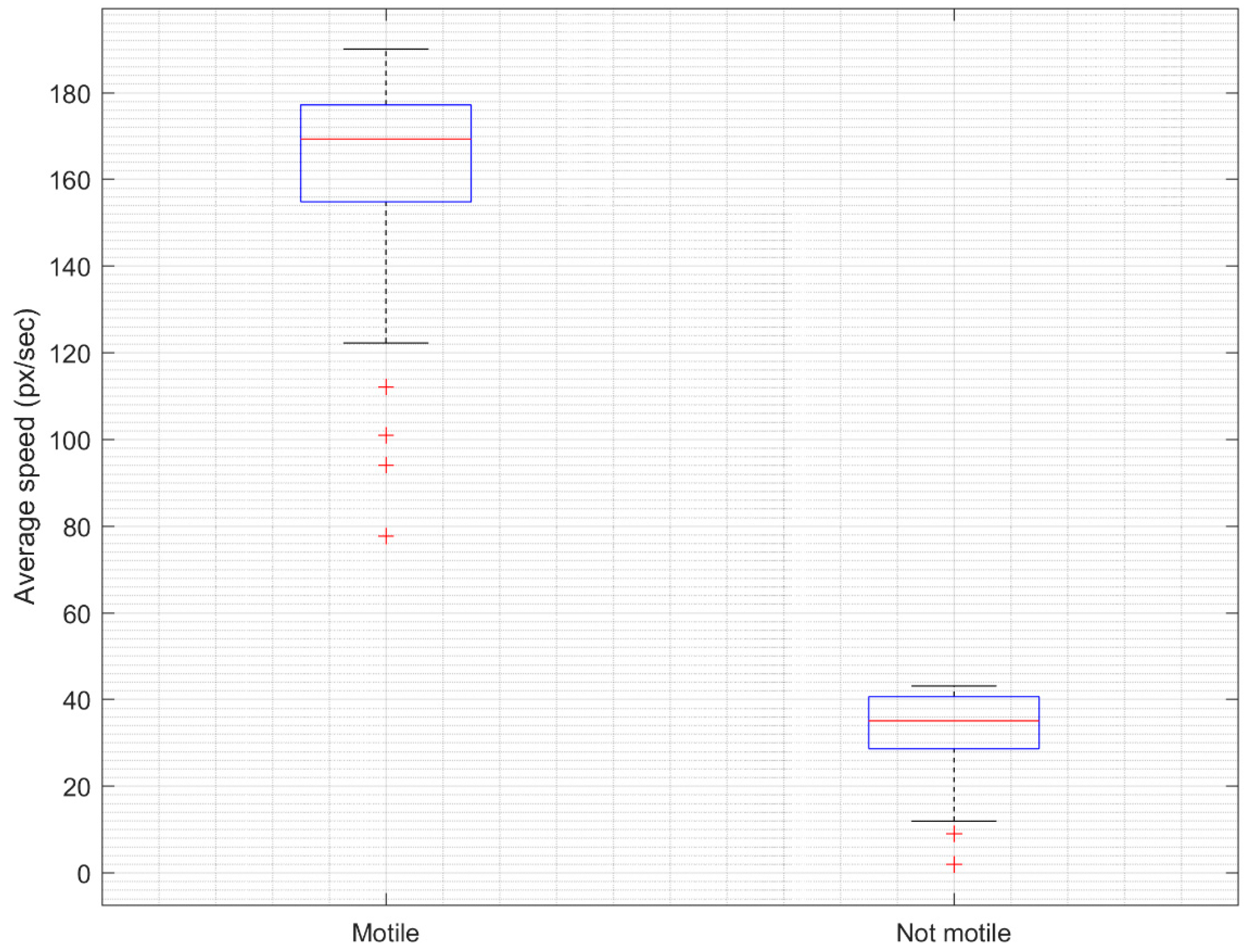
3.1. Results of Spermatozoid Viability Evaluation
3.2. Ablation Study
4. Evaluation and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, R.D.; Faure, N.; Glass, R.H. Artificial insemination of husband’s sperm. Fertil. Steril. 1979, 32, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Singh, A.K. Trends of male factor infertility, an important cause of infertility: A review of literature. J. Hum. Reprod. Sci. 2015, 8, 191–196. [Google Scholar] [CrossRef]
- Cooper, T.G.; Noonan, E.; von Eckardstein, S.; Auger, J.; Baker, H.W.G.; Behre, H.M.; Haugen, T.B.; Kruger, T.; Wang, C.; Mbizvo, M.T.; et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 2009, 16, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Guzick, D.S.; Overstreet, J.W.; Factor-Litvak, P.; Brazil, C.K.; Nakajima, S.T.; Coutifaris, C.; Vogel, D.L. Sperm morphology, motility, and concentration in fertile and infertile men. N. Engl. J. Med. 2001, 345, 1388–1393. [Google Scholar] [CrossRef]
- Wang, J.; Sauer, M.V. In vitro fertilization (IVF): A review of 3 decades of clinical innovation and technological advancement. Ther. Clin. Risk Manag. 2006, 2, 355–364. [Google Scholar] [CrossRef]
- Natali, I. Sperm Preparation Techniques for Artificial Insemination—Comparison of Sperm Washing, Swim Up, and Density Gradient Centrifugation Methods. In Artificial Insemination in Farm Animals; InTech: London, UK, 2011. [Google Scholar] [CrossRef]
- Koyun, E. The effect of intrauterine insemination time on semen parameters. J. Turk. Ger. Gynecol. Assoc. 2014, 15, 82–85. [Google Scholar] [CrossRef]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef]
- Sahlol, A.T.; Elaziz, M.A.; Jamal, A.T.; Damaševičius, R.; Hassan, O.F. A novel method for detection of tuberculosis in chest radiographs using artificial ecosystem-based optimisation of deep neural network features. Symmetry 2020, 12, 1146. [Google Scholar] [CrossRef]
- Orujov, F.; Maskeliūnas, R.; Damaševičius, R.; Wei, W. Fuzzy based image edge detection algorithm for blood vessel detection in retinal images. Appl. Soft Comput. J. 2020, 94, 106452. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, M.A.; Ahmed, F.; Mittal, M.; Goyal, L.M.; Jude Hemanth, D.; Satapathy, S.C. Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recognit. Lett. 2020, 131, 193–204. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, X.; Sun, S.; Wu, D.; Bai, J.; Yin, Y.; de Albuquerque, V.H.C. Learning physical properties in complex visual scenes: An intelligent machine for perceiving blood flow dynamics from static CT angiography imaging. Neural Netw. 2020, 123, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhou, B.; Połap, D.; Woźniak, M. A regional adaptive variational PDE model for computed tomography image reconstruction. Pattern Recognit. 2019, 92, 64–81. [Google Scholar] [CrossRef]
- Woźniak, M.; Połap, D.; Kośmider, L.; Cłapa, T. Automated fluorescence microscopy image analysis of pseudomonas aeruginosa bacteria in alive and dead stadium. Eng. Appl. Artif. Intell. 2018, 67, 100–110. [Google Scholar] [CrossRef]
- Ke, Q.; Zhang, J.; Wei, W.; Damaševičius, R.; Woźniak, M. Adaptive independent subspace analysis of brain magnetic resonance imaging data. IEEE Access 2019, 7, 12252–12261. [Google Scholar] [CrossRef]
- Fernandez, E.I.; Ferreira, A.S.; Cecílio, M.H.M.; Chéles, D.S.; de Souza, R.C.M.; Nogueira, M.F.G.; Rocha, J.C. Artificial intelligence in the IVF laboratory: Overview through the application of different types of algorithms for the classification of reproductive data. J. Assist. Reprod. Genet. 2020, 37, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Nissen, M.S.; Krause, O.; Almstrup, K.; Kjærulff, S.; Nielsen, T.T.; Nielsen, M. Convolutional Neural Networks for Segmentation and Object Detection of Human Semen. In Image Analysis. SCIA 2017. Lecture Notes in Computer Science; Sharma, P., Bianchi, F., Eds.; Springer: Cham, Switzerland, 2017; Volume 10269. [Google Scholar] [CrossRef]
- Hicks, S.A.; Andersen, J.M.; Witczak, O.; Thambawita, V.; Halvorsen, P.; Hammer, H.L.; Riegler, M.A. Machine learning-based analysis of sperm videos and participant data for male fertility prediction. Sci. Rep. 2019, 9, 16770. [Google Scholar] [CrossRef] [PubMed]
- Movahed, R.A.; Mohammadi, E.; Orooji, M. Automatic segmentation of sperm’s parts in microscopic images of human semen smears using concatenated learning approaches. Comput. Biol. Med. 2019, 109, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, K.K.; Afify, H.M.; Fouda, F.; Hassanien, A.E.; Bhattacharyya, S.; Vaclav, S. Classification of human sperm head in microscopic images using twin support vector machine and neural network. In Advances in Intelligent Systems and Computing; Springer: Singapore, 2020; pp. 857–871. [Google Scholar] [CrossRef]
- Thambawita, V.; Pål Halvorsen, P.; Hammer, H.; Riegler, M.; Haugen, T.B. Extracting temporal features into a spatial domain using autoencoders for sperm video analysis. arXiv 2019, arXiv:1911.03100. [Google Scholar]
- Butola, A.; Popova, D.; Prasad, D.K.; Ahmad, A.; Habib, A.; Tinguely, J.C.; Ahluwalia, B.S. High spatially sensitive quantitative phase imaging assisted with deep neural network for classification of human spermatozoa under stressed condition. Sci. Rep. 2020, 10, 13118. [Google Scholar] [CrossRef]
- Ilhan, H.O.; Serbes, G.; Aydin, N. Automated sperm morphology analysis approach using a directional masking technique. Comput. Biol. Med. 2020, 122, 103845. [Google Scholar] [CrossRef]
- Ilhan, H.O.; Sigirci, I.O.; Serbes, G.; Aydin, N. A fully automated hybrid human sperm detection and classification system based on mobile-net and the performance comparison with conventional methods. Med Biol. Eng. Comput. 2020, 58, 1047–1068. [Google Scholar] [CrossRef]
- Iqbal, I.; Mustafa, G.; Ma, J. Deep Learning-Based Morphological Classification of Human Sperm Heads. Diagnostics 2020, 10, 325. [Google Scholar] [CrossRef]
- Javadi, S.; Mirroshandel, S.A. A novel deep learning method for automatic assessment of human sperm images. Comput. Biol. Med. 2019, 109, 182–194. [Google Scholar] [CrossRef]
- Kandel, M.E.; Rubessa, M.; He, Y.R.; Schreiber, S.; Meyers, S.; Naves, L.M.; Popescu, G. Reproductive outcomes predicted by phase imaging with computational specificity of spermatozoon ultrastructure. Proc. Natl. Acad. Sci. USA 2020, 117, 18302–18309. [Google Scholar] [CrossRef]
- Riordon, J.; McCallum, C.; Sinton, D. Deep learning for the classification of human sperm. Comput. Biol. Med. 2019, 111, 103342. [Google Scholar] [CrossRef]
- Shaker, F.; Monadjemi, S.A.; Alirezaie, J.; Naghsh-Nilchi, A.R. A dictionary learning approach for human sperm heads classification. Comput. Biol. Med. 2017, 91, 181–190. [Google Scholar] [CrossRef]
- McCallum, C.; Riordon, J.; Wang, Y.; Kong, T.; You, J.B.; Sanner, S.; Lagunov, A.; Hannam, T.G.; Jarvi, K.; Sinton, S. Deep learning-based selection of human sperm with high DNA integrity. Commun. Biol. 2019, 2, 250. [Google Scholar] [CrossRef]
- Velasco, J.S.; Arago, N.M.; Mamba, R.M.; Padilla, M.V.C.; Ramos, J.P.M.; Virrey, G.C. Cattle sperm classification using transfer learning models. Int. J. Emerg. Trends Eng. Res. 2020, 8, 4325–4331. [Google Scholar] [CrossRef]
- Velasco, J.S.; Padilla, M.V.C.; Arago, N.M.; De Vera, E.P.E.; Domingo, F.E.M.; Ramos, R.E.R. Canine semen evaluation using transfer learning models. Int. J. Emerg. Trends Eng. Res. 2020, 8, 1–6. [Google Scholar]
- Hidayatullah, P.; Wang, X.; Yamasaki, T.; Mengko, T.L.E.R.; Munir, R.; Barlian, A.; Sukmawati, E.; Supraptono, S. DeepSperm: A robust and real-time bull sperm-cell detection in densely populated semen videos. arXiv 2020, arXiv:2003.01395. [Google Scholar]
- Hidayatullah, P.; Mengko, T.E.R.; Munir, R.; Barlian, A. A semiautomatic sperm cell data annotator for convolutional neural network. In Proceedings of the 5th International Conference on Science in Information Technology: Embracing Industry 4.0: Towards Innovation in Cyber Physical System, ICSITech 2019, Yogyakarta, Indonesia, 23–24 October 2019; pp. 211–216. [Google Scholar] [CrossRef]
- Haugen, T.B.; Hicks, S.A.; Andersen, J.M.; Witczak, O.; Hammer, H.L.; Borgli, R.; Halvorsen, P.; Riegler, M. VISEM: A multimodal video dataset of human spermatozoa. In Proceedings of the 10th ACM Multimedia Systems Conference, MMSys’19, Amherst, MA, USA, 18–21 June 2019; pp. 261–266. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Alom, Z.; Hasan, M.; Yakopcic, C.; Tarek, M.; Taha, T.M. Inception Recurrent Convolutional Neural Network for Object Recognition. arXiv 2017, arXiv:1704.07709. [Google Scholar]
- Holt, W.V.; Moore, H.D.M.; Hillier, S.G. Computer-assisted measurement of sperm swimming speed in human semen: Correlation of results with in vitro fertilization assays. Fertil. Steril. 1985, 44, 112–119. [Google Scholar] [CrossRef]
- Shaker, F. Human Sperm Head Morphology Dataset (HuSHeM) [Data set]; Mendeley: London, UK, 2018. [Google Scholar] [CrossRef]











| Parameter | Statistics | Value |
|---|---|---|
| Sperm concentration (×106/mL) | Median (range) Mean ± std | 68 (4–350) 82.33 ± 64.11 |
| Average number of sperm heads in one frame | Median (range) | 34 (2–175) |
| Mean ± std | 41.16 ± 32.05 |
| No. | Frame ID | Objects, Pcs | Experimental Viability, % | Estimated Viability, % | Deviation, % | ||
|---|---|---|---|---|---|---|---|
| Found | Live | Viable | |||||
| 1 | 1 | 80 | 78 | 68 | 85 | 87 | 2 |
| 2 | 14 | 6 | 5 | 4 | 96 | 84 | 12 |
| 3 | 22 | 21 | 15 | 10 | 75 | 72 | 3 |
| 4 | 23 | 10 | 7 | 6 | 85 | 85 | 0 |
| 5 | 30 | 21 | 20 | 11 | 91 | 87 | 4 |
| 6 | 42 | 55 | 52 | 43 | 85 | 83 | 2 |
| 7 | 51 | 90 | 88 | 77 | 86 | 88 | 2 |
| 8 | 79 | 92 | 86 | 76 | 97 | 88 | 9 |
| 9 | 81 | 80 | 74 | 64 | 81 | 87 | 6 |
| 10 | 82 | 39 | 34 | 24 | 85 | 72 | 13 |
| Learning Speed | Number of Steps | Objects | Average Score | ||
|---|---|---|---|---|---|
| Mark | Recognize | Wrong | |||
| 0.00002 | 2000 | 10 | 7 | 3 | 0.71 |
| 0.0002 | 2000 | 30 | 30 | 0 | 0.85 |
| 0.002 | 2000 | 87 | 30 | 0 | 0.78 |
| Number of Iterations | Detected Objects | Identified Objects | Unrecognized Objects | Accuracy | Average Score |
|---|---|---|---|---|---|
| 50,000 | 30 | 30 | 3 | 0.9 | 0.88 |
| 100,000 | 32 | 32 | 1 | 0.97 | 0.86 |
| 150,000 | 31 | 31 | 2 | 0.94 | 0.96 |
| 200,000 | 33 | 33 | 0 | 1 | 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valiuškaitė, V.; Raudonis, V.; Maskeliūnas, R.; Damaševičius, R.; Krilavičius, T. Deep Learning Based Evaluation of Spermatozoid Motility for Artificial Insemination. Sensors 2021, 21, 72. https://doi.org/10.3390/s21010072
Valiuškaitė V, Raudonis V, Maskeliūnas R, Damaševičius R, Krilavičius T. Deep Learning Based Evaluation of Spermatozoid Motility for Artificial Insemination. Sensors. 2021; 21(1):72. https://doi.org/10.3390/s21010072
Chicago/Turabian StyleValiuškaitė, Viktorija, Vidas Raudonis, Rytis Maskeliūnas, Robertas Damaševičius, and Tomas Krilavičius. 2021. "Deep Learning Based Evaluation of Spermatozoid Motility for Artificial Insemination" Sensors 21, no. 1: 72. https://doi.org/10.3390/s21010072
APA StyleValiuškaitė, V., Raudonis, V., Maskeliūnas, R., Damaševičius, R., & Krilavičius, T. (2021). Deep Learning Based Evaluation of Spermatozoid Motility for Artificial Insemination. Sensors, 21(1), 72. https://doi.org/10.3390/s21010072






