Affective Computing on Machine Learning-Based Emotion Recognition Using a Self-Made EEG Device
Abstract
:1. Introduction
- We proposed a novel end-to-end affective computing based on machine learning for emotion recognition utilizing a wireless and wearable custom-designed EEG device with a low-cost and compact design.
- The combination of sample entropy feature extraction and 1D-CNN was introduced and achieved the best performance among the proposed entropy measures and machine learning models for two kinds of EEG emotion recognition experiments, including the subject-dependent and subject-independent cases.
- We also investigated that T8 in the temporal lobe was adopted most frequently through the electrode selection method, implying that this position would have further valuable information than other locations for emotion recognition.
2. Materials and Methods
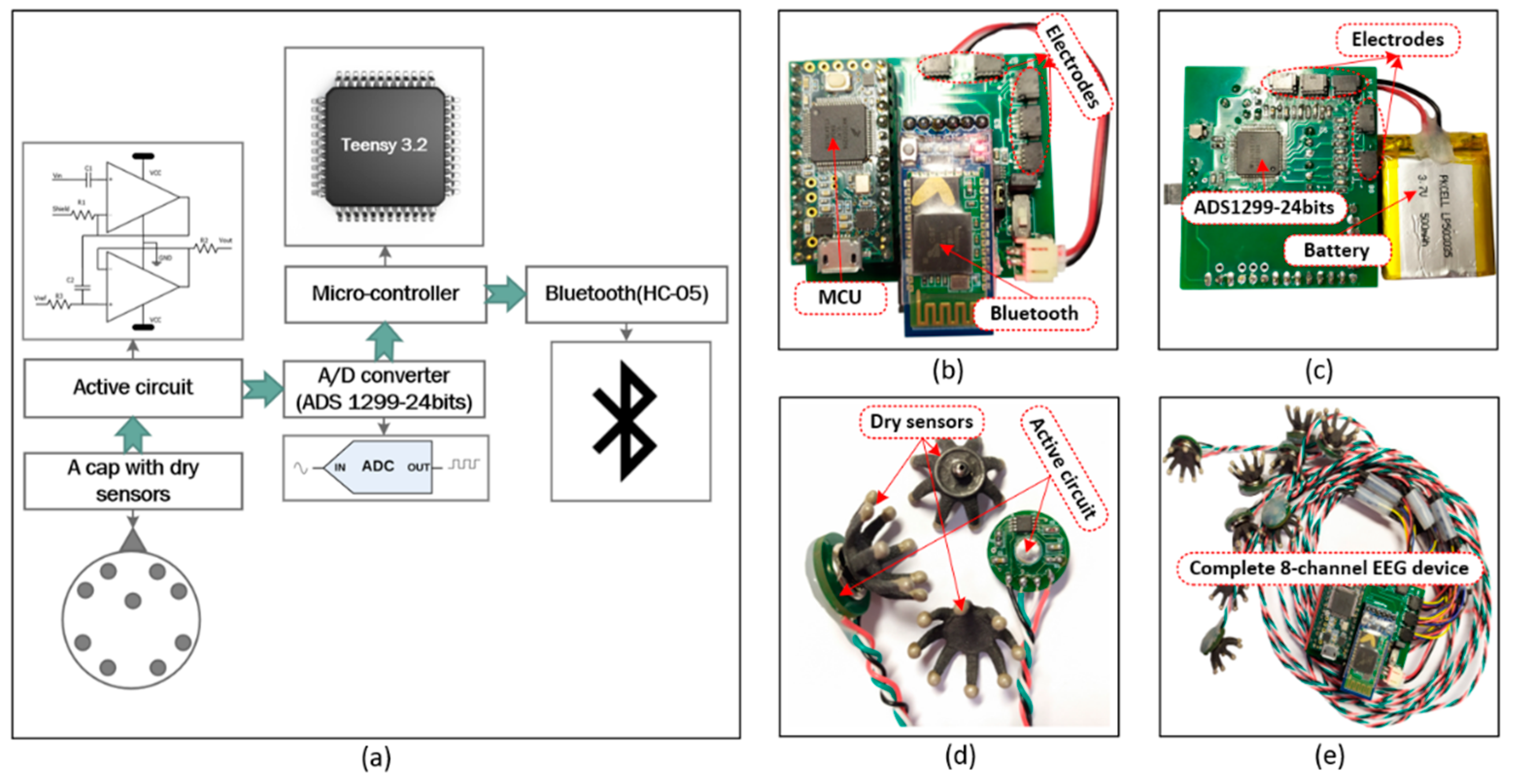
2.1. Eight-Channel EEG Recording
2.2. Stimulus and Protocol
2.3. Feature Extraction
2.3.1. Permutation Entropy (PEE)
2.3.2. Singular Value Decomposition Entropy (SVE)
2.3.3. Approximate Entropy (APE)
2.3.4. Sample Entropy (SAE)
2.3.5. Spectral Entropy (SPE)
2.3.6. Continuous Wavelet Transform Entropy (CWE)
2.4. Electrode Selection
2.5. Dimensionality Reduction
2.6. Classification
2.6.1. Support Vector Machine (SVM)
2.6.2. Multi-Layer Perceptron (MLP)
2.6.3. 1-D Convolutional Neural Neworks (1D-CNN)
3. Results
3.1. EEG Features
3.2. EEG Electrode Selection and Rejection
3.3. Classification Results
3.4. Subject-Dependent and Subject-Independent
3.5. Dimensionality Reduction and Data Visualization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, W.-L.; Zhu, J.-Y.; Lu, B.-L. Identifying Stable Patterns over Time for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2017, 10, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Stets, J.E.; Turner, J.H. (Eds.) Handbook of the Sociology of Emotions: Volume II; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Picard, R.W. Affective Computing; MIT Press: Cambridge, MA, USA, 1997. [Google Scholar]
- Zhou, Q. Multi-layer affective computing model based on emotional psychology. Electron. Commer. Res. 2017, 18, 109–124. [Google Scholar] [CrossRef]
- Tao, J.; Tan, T. Affective computing: A review. In Proceedings of the International Conference on Affective Computing and Intelligent Interaction, Beijing, China, 22–24 October 2005; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Paschen, J.; Kietzmann, J.; Kietzmann, T.C. Artificial intelligence (AI) and its implications for market knowledge in B2B marketing. J. Bus. Ind. Mark. 2019, 34, 1410–1419. [Google Scholar] [CrossRef]
- Nguyen, T.-H.; Chung, W.-Y. Negative News Recognition during Social Media News Consumption Using EEG. IEEE Access 2019, 7, 133227–133236. [Google Scholar] [CrossRef]
- Krishna, N.M.; Sekaran, K.; Vamsi, A.V.N.; Ghantasala, G.P.; Chandana, P.; Kadry, S. An efficient mixture model approach in brain-machine interface systems for extracting the psychological status of mentally impaired persons using EEG signals. IEEE Access 2019, 7, 77905–77914. [Google Scholar] [CrossRef]
- Šalkevicius, J.; Damaševičius, R.; Maskeliunas, R.; Laukienė, I. Anxiety Level Recognition for Virtual Reality Therapy System Using Physiological Signals. Electronics 2019, 8, 1039. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K. Remarks on SVM-based emotion recognition from multi-modal bio-potential signals. In Proceedings of the 13th IEEE International Workshop on Robot and Human Interactive Communication, Okayama, Japan, 20–22 September 2004. [Google Scholar]
- Quintana, D.S.; Guastella, A.J.; Outhred, T.; Hickie, I.; Kemp, A.H. Heart rate variability is associated with emotion recognition: Direct evidence for a relationship between the autonomic nervous system and social cognition. Int. J. Psychophysiol. 2012, 86, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Goshvarpour, A.; Abbasi, A.; Goshvarpour, A. An accurate emotion recognition system using ECGand GSR signals and matching pursuit method. Biomed. J. 2017, 40, 355–368. [Google Scholar] [CrossRef]
- Chanel, G.; Kronegg, J.; Grandjean, D.; Pun, T. Emotion assessment: Arousal evaluation using EEG’s and peripheral phys-iological signals. In International Workshop on Multimedia Content Representation, Classification and Security; Springer: Berlin, Germany, 2006. [Google Scholar]
- Liu, Z.; Wang, S. Emotion recognition using hidden Markov models from facial temperature sequence. In Proceedings of the International Conference on Affective Computing and Intelligent Interaction, Memphis, TN, USA, 9–12 October 2011; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Ahern, G.L.; Schwartz, G.E. Differential lateralization for positive and negative emotion in the human brain: EEG spectral analysis. Neuropsychologia 1985, 23, 745–755. [Google Scholar] [CrossRef]
- Ball, T.; Kern, M.; Mutschler, I.; Aertsen, A.; Schulze-Bonhage, A. Signal quality of simultaneously recorded invasive and non-invasive EEG. NeuroImage 2009, 46, 708–716. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-P.; Wang, C.-H.; Jung, T.-P.; Wu, T.-L.; Jeng, S.-K.; Duann, J.-R.; Chen, J.-H. EEG-Based Emotion Recognition in Music Listening. IEEE Trans. Biomed. Eng. 2010, 57, 1798–1806. [Google Scholar] [PubMed]
- Tandle, A.; Jog, N.; D’cunha, P.; Chheta, M. Classification of artefacts in EEG signal recordings and overview of removing techniques. Int. J. Comput. Appl. 2015, 975, 8887. [Google Scholar]
- Zhang, X.D. Entropy for the Complexity of Physiological Signal Dynamics. Healthc. Big Data Manag. 2017, 1028, 39–53. [Google Scholar]
- De Luca, A.; Termini, S. A definition of a nonprobabilistic entropy in the setting of fuzzy sets theory. Inf. Control 1972, 20, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Jenke, R.; Peer, A.; Buss, M. Feature Extraction and Selection for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2014, 5, 327–339. [Google Scholar] [CrossRef]
- Alotaiby, T.; Abd El-Samie, F.E.; Alshebeili, S.A.; Ahmad, I. A review of channel selection algorithms for EEG signal processing. EURASIP J. Adv. Signal Process. 2015, 2015, 66. [Google Scholar] [CrossRef] [Green Version]
- St, L.; Wold, S. Analysis of variance (ANOVA). Chemom. Intell. Lab. Syst. 1989, 6, 259–272. [Google Scholar]
- Abdi, H.; Williams, L.J. Tukey’s honestly significant difference (HSD) test. Encycl. Res. Des. 2010, 3, 1–5. [Google Scholar]
- Dry Sensor. 2016. Available online: https://www.cgxsystems.com/quick-30 (accessed on 21 May 2021).
- Texas Instruments. ADS1299 Low-Noise, 8-Channel, 24-Bit, Analog-to-Digital Converter for EEG and Biopotential Measurements; Data Sheet; Texas Instruments: Dallas, TX, USA, 2017. [Google Scholar]
- Zheng, W.L.; Lu, B.L. Investiating Critical Frequency Bands and Channels for EEG-Based Emotion Recognition. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Balconi, M.; Brambilla, E.; Falbo, L. Appetitive vs. defensive responses to emotional cues. Autonomic measures and brain oscillation modulation. Brain Res. 2009, 1296, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Rolls, E.; Hornak, J.; Wade, D.; McGrath, J. Emotion-related learning in patients with social and emotional changes associated with frontal lobe damage. J. Neurol. Neurosurg. Psychiatry 1994, 57, 1518–1524. [Google Scholar] [CrossRef]
- Homan, R.W.; Herman, J.; Purdy, P. Cerebral location of international 10–20 system electrode placement. Electroencephalogr. Clin. Neurophysiol. 1987, 66, 376–382. [Google Scholar] [CrossRef]
- Maskeliunas, R.; Damasevicius, R.; Martisius, I.; Vasiljevas, M. Consumer grade EEG devices: Are they usable for control tasks? PeerJ 2016, 4, e1746. [Google Scholar] [CrossRef] [PubMed]
- Britton, J.C.; Taylor, S.; Sudheimer, K.D.; Liberzon, I. Facial expressions and complex IAPS pictures: Common and differential networks. NeuroImage 2006, 31, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A database for amotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Dan-Glauser, E.S.; Scherer, K.R. The Geneva affective picture database (GAPED): A new 730-picture database focusing on valence and normative significance. Behav. Res. Methods 2011, 43, 468–477. [Google Scholar] [CrossRef]
- Marchewka, A.; Żurawski, Ł.; Jednoróg, K.; Grabowska, A. The Nencki Affective Picture System (NAPS): Introduction to a novel, standardized, wide-range, high-quality, realistic picture database. Behav. Res. Methods 2014, 46, 596–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carretié, L.; Tapia, M.; López-Martín, S.; Albert, J. EmoMadrid: An emotional pictures database for affect research. Motiv. Emot. 2019, 43, 929–939. [Google Scholar] [CrossRef]
- Ellard, K.K.; Farchione, T.J.; Barlow, D.H. Relative Effectiveness of Emotion Induction Procedures and the Role of Personal Relevance in a Clinical Sample: A Comparison of Film, Images, and Music. J. Psychopathol. Behav. Assess. 2011, 34, 232–243. [Google Scholar] [CrossRef]
- Choi, J.; Rangan, P.; Singh, S.N. Do Cold Images Cause Cold-Heartedness? The Impact of Visual Stimuli on the Effectiveness of Negative Emotional Charity Appeals. J. Advert. 2016, 45, 417–426. [Google Scholar] [CrossRef]
- Mikels, J.A.; Fredrickson, B.L.; Samanez-Larkin, G.; Lindberg, C.M.; Maglio, S.J.; Reuter-Lorenz, P.A. Emotional category data on images from the international affective picture system. Behav. Res. Methods 2005, 37, 626–630. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Zheng, W.-L.; Lu, B.-L. Cross-subject and cross-gender emotion classification from EEG. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering, Toronto, ON, Canada, 7–12 June 2015; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Tolegenova, A.A.; Kustubayeva, A.M.; Matthews, G. Trait meta-mood, gender and EEG response during emotion-regulation. Personal. Individ. Differ. 2014, 65, 75–80. [Google Scholar] [CrossRef]
- Ouyang, G.; Li, J.; Liu, X.; Li, X. Dynamic characteristics of absence EEG recordings with multiscale permutation entropy analysis. Epilepsy Res. 2013, 104, 246–252. [Google Scholar] [CrossRef]
- Prabhakar, S.K.; Rajaguru, H. Performance comparison of fuzzy mutual information as dimensionality reduction techniques and SRC, SVD and approximate entropy as post classifiers for the classification of epilepsy risk levels from EEG signals. In Proceedings of the 2015 IEEE Student Symposium in Biomedical Engineering & Sciences (ISSBES), Shah Alam, Malaysia, , 4 November 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar]
- Srinivasan, V.; Eswaran, C.; Sriraam, N. Approximate Entropy-Based Epileptic EEG Detection Using Artificial Neural Networks. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 288–295. [Google Scholar] [CrossRef]
- Jie, X.; Cao, R.; Li, L. Emotion recognition based on the sample entropy of EEG. Bio-Med. Mater. Eng. 2014, 24, 1185–1192. [Google Scholar] [CrossRef]
- Seitsonen, E.R.J.; Korhonen, I.; van Gils, M.; Huiku, M.; Lötjönen, J.M.P.; Korttila, K.T.; Yli-Hankala, A.M. EEG spectral entropy, heart rate, photoplethysmography and motor responses to skin incision during sevoflurane anaesthesia. Acta Anaesthesiol. Scand. 2005, 49, 284–292. [Google Scholar] [CrossRef]
- Adeli, H.; Zhou, Z.; Dadmehr, N. Analysis of EEG records in an epileptic patient using wavelet transform. J. Neurosci. Methods 2003, 123, 69–87. [Google Scholar] [CrossRef]
- Gannouni, S.; Aledaily, A.; Belwafi, K.; Aboalsamh, H. Emotion detection using electroencephalography signals and a zero-time windowing-based epoch estimation and relevant electrode identification. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Isajiw, W.W.; Sev’er, A.; Driedger, L. Ethnic Identity and Social Mobility: A Test of the ‘Drawback Model’. Can. J. Sociol. Cah. Can. Sociol. 1993, 18, 177–196. [Google Scholar] [CrossRef]
- Ringnér, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef]
- Belkina, A.C.; Ciccolella, C.O.; Anno, R.; Halpert, R.; Spidlen, J.; Snyder-Cappione, J.E. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parra-Hernández, R.M.; Posada-Quintero, J.I.; Acevedo-Charry, O.; Posada-Quintero, H.F. Uniform Manifold Approximation and Projection for Clustering Taxa through Vocalizations in a Neotropical Passerine (Rough-Legged Tyrannulet, Phyllomyias burmeisteri). Animals 2020, 10, 1406. [Google Scholar] [CrossRef]
- Wang, L. (Ed.) Support Vector Machines: Theory and Applications; Springer Science & Business Media: Chicago, IL, USA, 2005; Volume 177. [Google Scholar]
- Zare, M.; Pourghasemi, H.R.; Vafakhah, M.; Pradhan, B. Landslide susceptibility mapping at Vaz Watershed (Iran) using an artificial neural network model: A comparison between multilayer perceptron (MLP) and radial basic function (RBF) algorithms. Arab. J. Geosci. 2012, 6, 2873–2888. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Streiner, D.; Cairney, J. What’s under the ROC? An introduction to receiver operating characteristics curves. Can. J. Psychiatry 2007, 52, 121–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]













| Electrode | p-Value (ANOVA and Tukey’s HSD Test) | Decision (p ≤ 0.05) | ||
|---|---|---|---|---|
| neg-neu | neg-pos | neu-pos | ||
| 1 | 0.001 | 0.8130 | 0.001 | reject |
| 2 | 0.001 | 0.001 | 0.001 | adopt |
| 3 | 0.5900 | 0.0365 | 0.2916 | reject |
| 4 | 0.001 | 0.001 | 0.001 | adopt |
| 5 | 0.1053 | 0.001 | 0.001 | reject |
| 6 | 0.001 | 0.001 | 0.001 | adopt |
| 7 | 0.0029 | 0.001 | 0.8367 | reject |
| 8 | 0.0225 | 0.001 | 0.001 | adopt |
| Subject | Selected Electrodes | |||||
|---|---|---|---|---|---|---|
| PEE | SVE | APE | SAE | SPE | CWE | |
| 1 | 1, 3, 4, 5, 6, 7, 8 | 6, 7 | 2, 4, 5, 6, 8 | 4, 5, 6, 7, 8 | 1, 2, 3, 6, 7 | 1, 2, 6, 7 |
| 2 | 2, 5 | 1, 3, 4, 5, 8 | 2, 4, 5, 8 | 2, 4, 6, 8 | 2, 4, 5, 6, 8 | 1, 2, 4, 5, 6 |
| 3 | 4, 8 | All electrodes | 3, 4, 6 | 1, 2, 3, 4, 6, 7, 8 | 1, 2, 3, 4, 7, 8 | 2, 4, 6, 7 |
| 4 | 6, 7 | 1, 6 | 4, 6 | 1, 7 | 1, 3, 6 | 1, 2, 6 |
| 5 | 1, 3, 4, 5, 6, 7, 8 | 2, 4, 6 | 2, 4, 6, 7 | 2, 3, 4, 6, 7 | 1, 2, 5 | 6, 8 |
| 6 | 5, 6, 7 | 1, 2, 4, 5, 6, 7, 8 | 4, 6 | 1, 2, 4, 5, 6, 7, 8 | 1, 2, 5, 6, 7, 8 | 1, 3, 6, 8 |
| 7 | 4, 8 | 1, 2, 4, 5, 6, 7, 8 | 7, 8 | 3, 4, 7, 8 | 7, 8 | 1, 4, 6 |
| 8 | 2, 5, 6 | 1, 3, 7 | 1, 7 | 1, 2, 7, 8 | 1, 7 | 1, 2, 5 |
| Subject | EEG Features | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PEE | SVE | APE | |||||||
| SVM (%) | MLP (%) | 1D-CNN (%) | SVM (%) | MLP (%) | 1D-CNN (%) | SVM (%) | MLP (%) | 1D-CNN (%) | |
| 1 | 83.33 | 70.22 | 79.64 | 73.14 | 74.14 | 81.97 | 69.17 | 78.33 | 75.88 |
| 2 | 85.93 | 76.54 | 83.46 | 81.87 | 70.28 | 77.78 | 74.92 | 72.59 | 75.39 |
| 3 | 69.45 | 67.92 | 69.44 | 87.52 | 89.72 | 90.35 | 65.89 | 65.83 | 66.58 |
| 4 | 68.57 | 70.04 | 70.65 | 72.99 | 71.03 | 82.22 | 67.67 | 68.22 | 68.56 |
| 5 | 75.92 | 75.65 | 80.98 | 81.58 | 78.17 | 84.05 | 74.27 | 75.56 | 76.25 |
| 6 | 70.54 | 71.39 | 71.27 | 81.25 | 79.72 | 84.62 | 66.95 | 67.84 | 68.01 |
| 7 | 71.75 | 72.45 | 71.55 | 79.49 | 79.04 | 83.70 | 63.47 | 65.48 | 69.16 |
| 8 | 68.98 | 71.13 | 70.42 | 72.67 | 79.80 | 84.44 | 68.50 | 69.57 | 68.09 |
| Mean | 74.31 | 71.92 | 74.68 | 78.81 | 77.74 | 83.64 | 68.85 | 70.43 | 70.99 |
| Std | 6.73 | 2.71 | 5.30 | 5.04 | 5.78 | 3.28 | 3.69 | 4.35 | 3.82 |
| Subject | EEG Features | ||||||||
| SAE | SPE | CWE | |||||||
| SVM (%) | MLP (%) | 1D-CNN (%) | SVM (%) | MLP (%) | 1D-CNN (%) | SVM (%) | MLP (%) | 1D-CNN (%) | |
| 1 | 70.89 | 80.95 | 86.75 | 78.38 | 75.45 | 86.12 | 68.72 | 71.63 | 72.18 |
| 2 | 87.79 | 92.90 | 93.89 | 87.50 | 84.72 | 84.90 | 73.29 | 72.61 | 72.37 |
| 3 | 83.70 | 81.28 | 86.26 | 79.97 | 84.98 | 80.56 | 66.92 | 65.95 | 66.88 |
| 4 | 73.61 | 81.83 | 83.06 | 79.82 | 82.57 | 81.50 | 64.74 | 65.52 | 66.76 |
| 5 | 84.06 | 80.81 | 84.75 | 73.10 | 77.17 | 76.26 | 75.23 | 75.63 | 76.55 |
| 6 | 79.33 | 79.76 | 85.94 | 72.59 | 74.44 | 77.59 | 68.56 | 67.93 | 68.25 |
| 7 | 78.19 | 75.65 | 82.17 | 74.57 | 77.22 | 76.99 | 65.62 | 66.84 | 67.87 |
| 8 | 77.32 | 78.97 | 83.68 | 75.93 | 80.05 | 80.78 | 69.54 | 70.97 | 72.91 |
| Mean | 79.36 | 81.52 | 85.81 | 77.73 | 79.58 | 80.59 | 69.08 | 69.64 | 70.47 |
| Std | 5.27 | 4.67 | 3.41 | 4.56 | 3.87 | 3.37 | 3.39 | 3.38 | 3.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mai, N.-D.; Lee, B.-G.; Chung, W.-Y. Affective Computing on Machine Learning-Based Emotion Recognition Using a Self-Made EEG Device. Sensors 2021, 21, 5135. https://doi.org/10.3390/s21155135
Mai N-D, Lee B-G, Chung W-Y. Affective Computing on Machine Learning-Based Emotion Recognition Using a Self-Made EEG Device. Sensors. 2021; 21(15):5135. https://doi.org/10.3390/s21155135
Chicago/Turabian StyleMai, Ngoc-Dau, Boon-Giin Lee, and Wan-Young Chung. 2021. "Affective Computing on Machine Learning-Based Emotion Recognition Using a Self-Made EEG Device" Sensors 21, no. 15: 5135. https://doi.org/10.3390/s21155135
APA StyleMai, N.-D., Lee, B.-G., & Chung, W.-Y. (2021). Affective Computing on Machine Learning-Based Emotion Recognition Using a Self-Made EEG Device. Sensors, 21(15), 5135. https://doi.org/10.3390/s21155135








