Development and Modification of Pre-miRNAs with a FRET Dye Pair for the Intracellular Visualization of Processing Intermediates That Are Generated in Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Syntheses of the Modified RNAs
2.2. Preparation of the Modified Pre-miRNAs
2.3. Measurements of UV–VIS Spectra and Tm
2.4. Fluorescent Spectroscopic Measurements
2.5. Dicing Assay
2.6. Cell Culture
2.7. Luciferase Assay
2.8. Fluorescent Imaging Analyses of the Pre-miRNA Transfected HeLa Cells
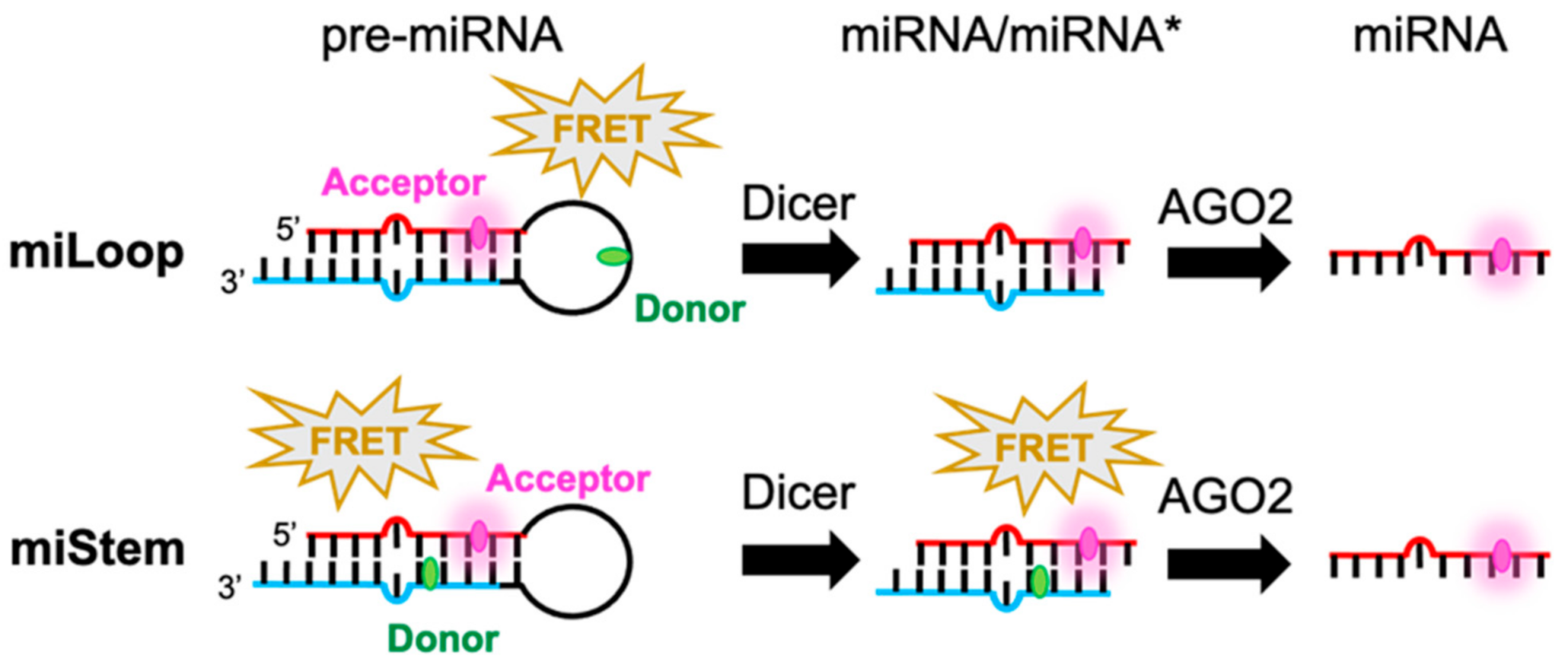
3. Results
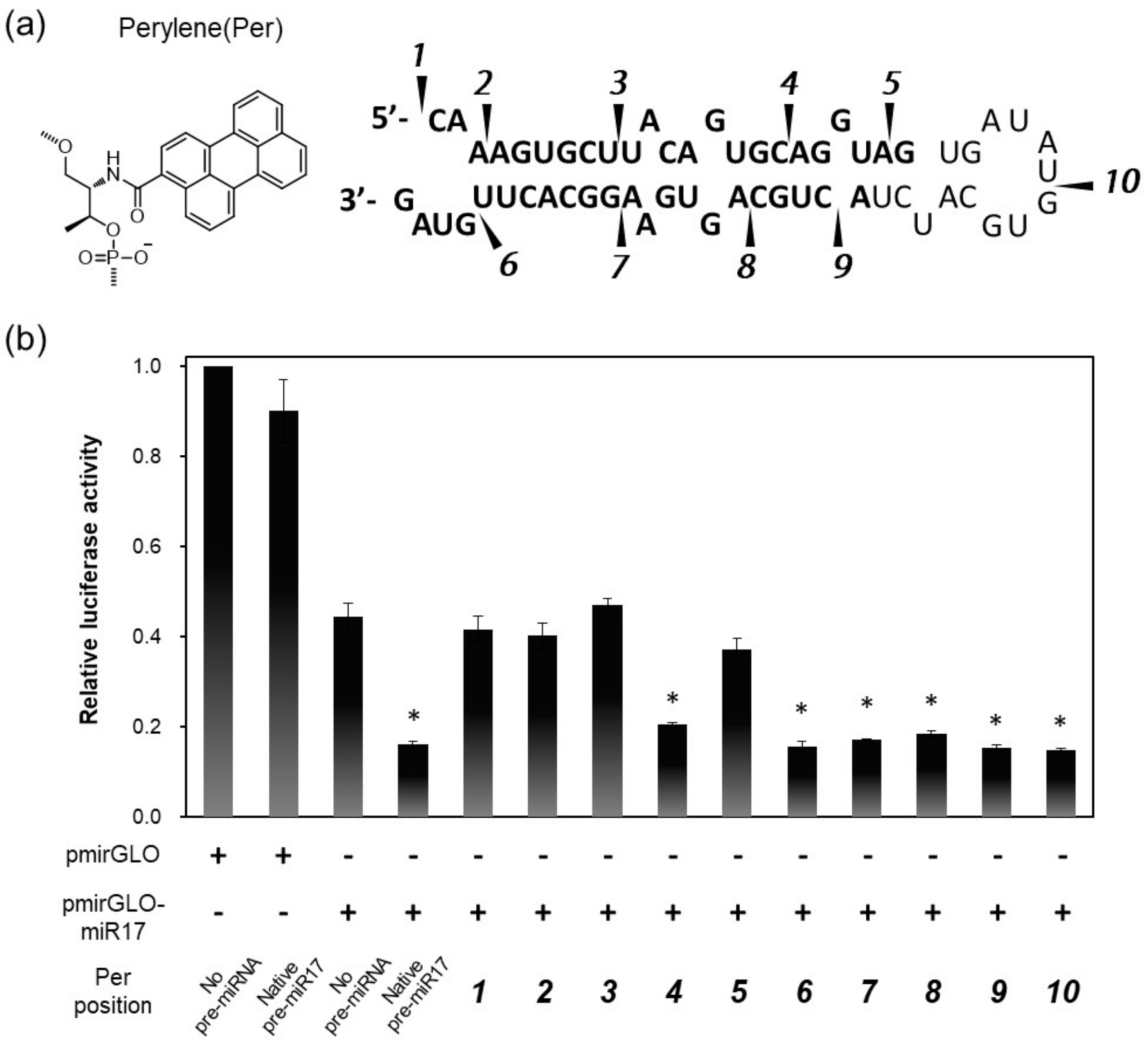
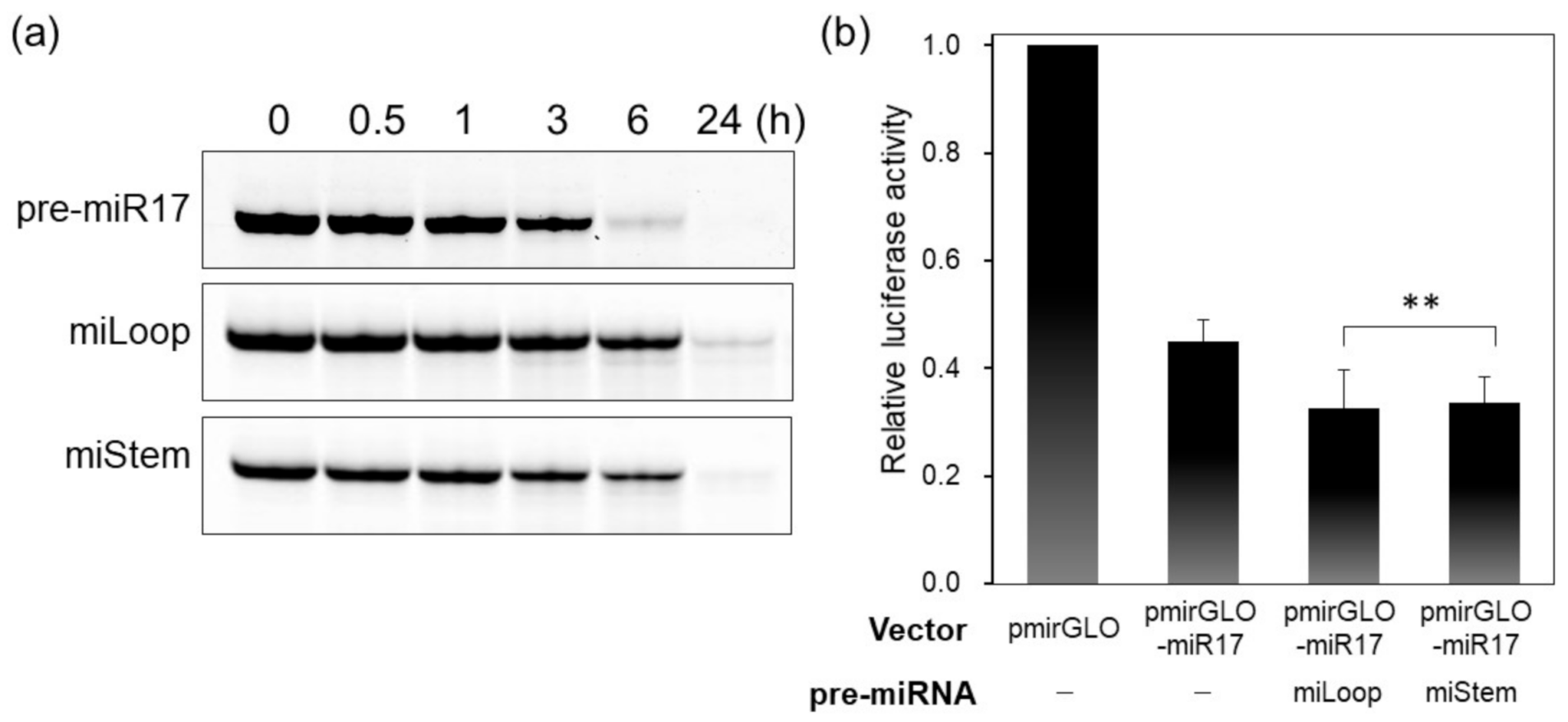
3.1. Activities of Pre-miR17s That Were Modified with the Fluorophore at Various Positions
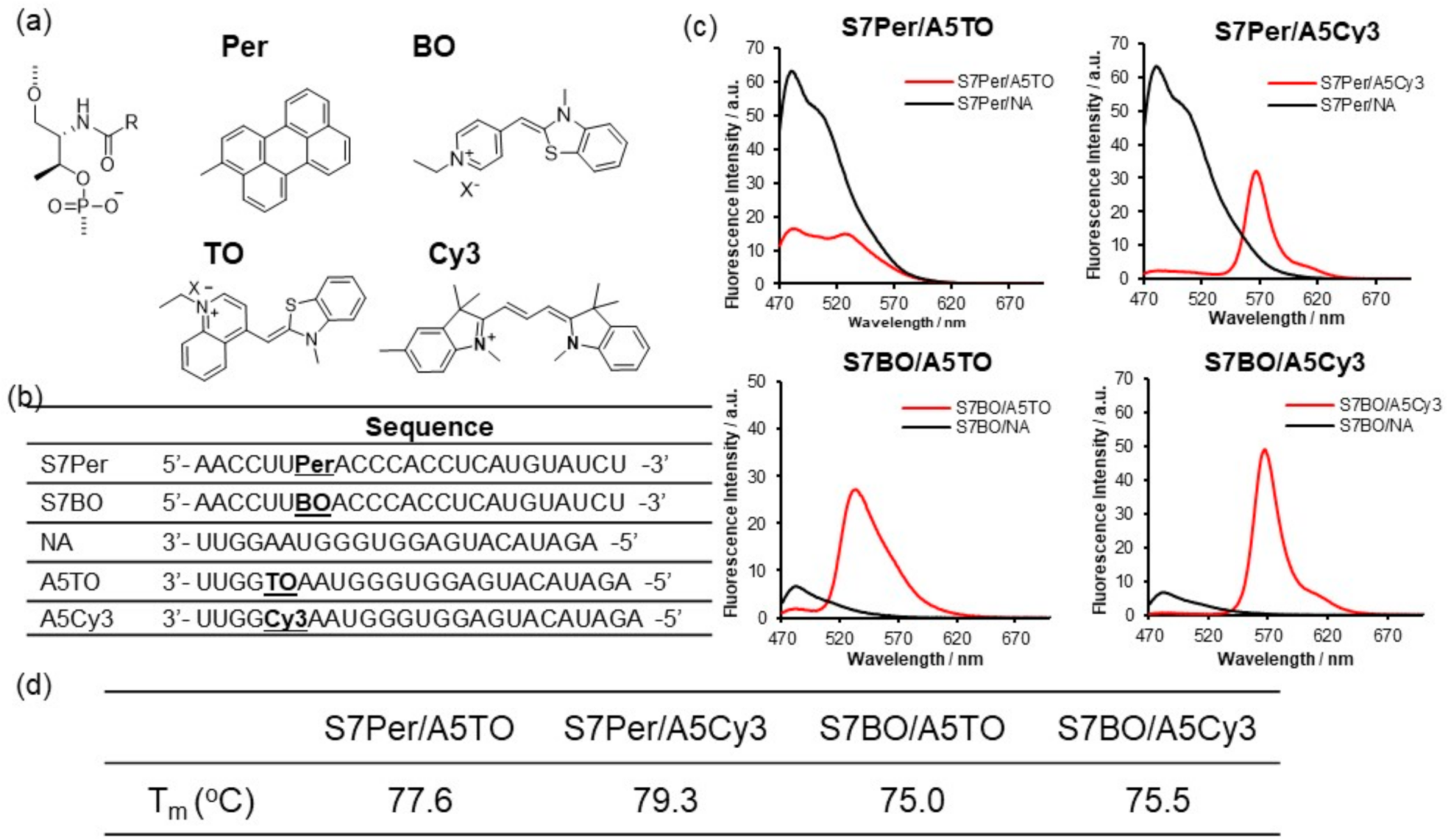
3.2. Investigation of Suitable FRET Dyes That Were Introduced into Pre-miR17
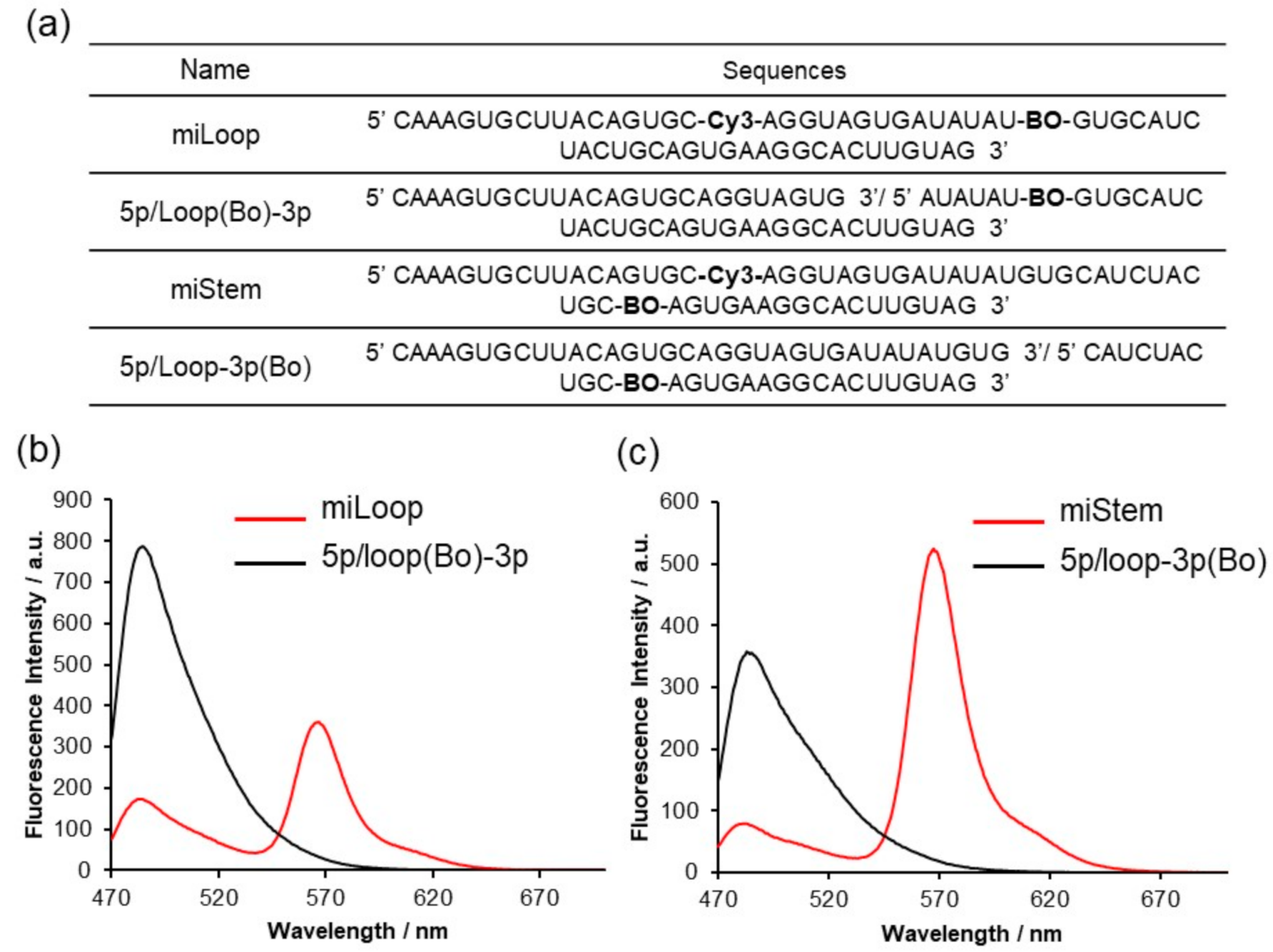
3.3. Biological Activities of the BO–Cy3 Pair-Modified Pre-miR17s
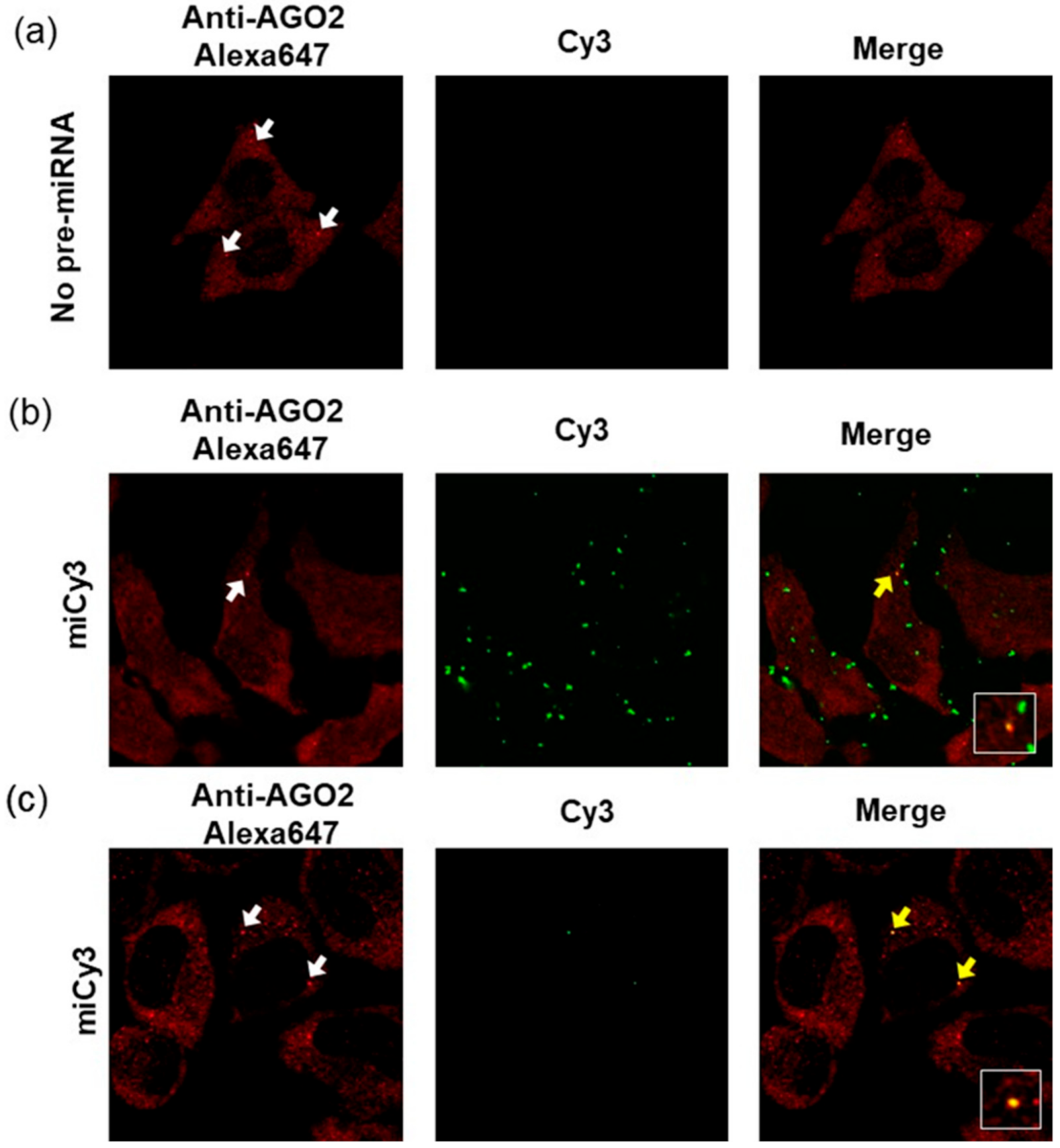
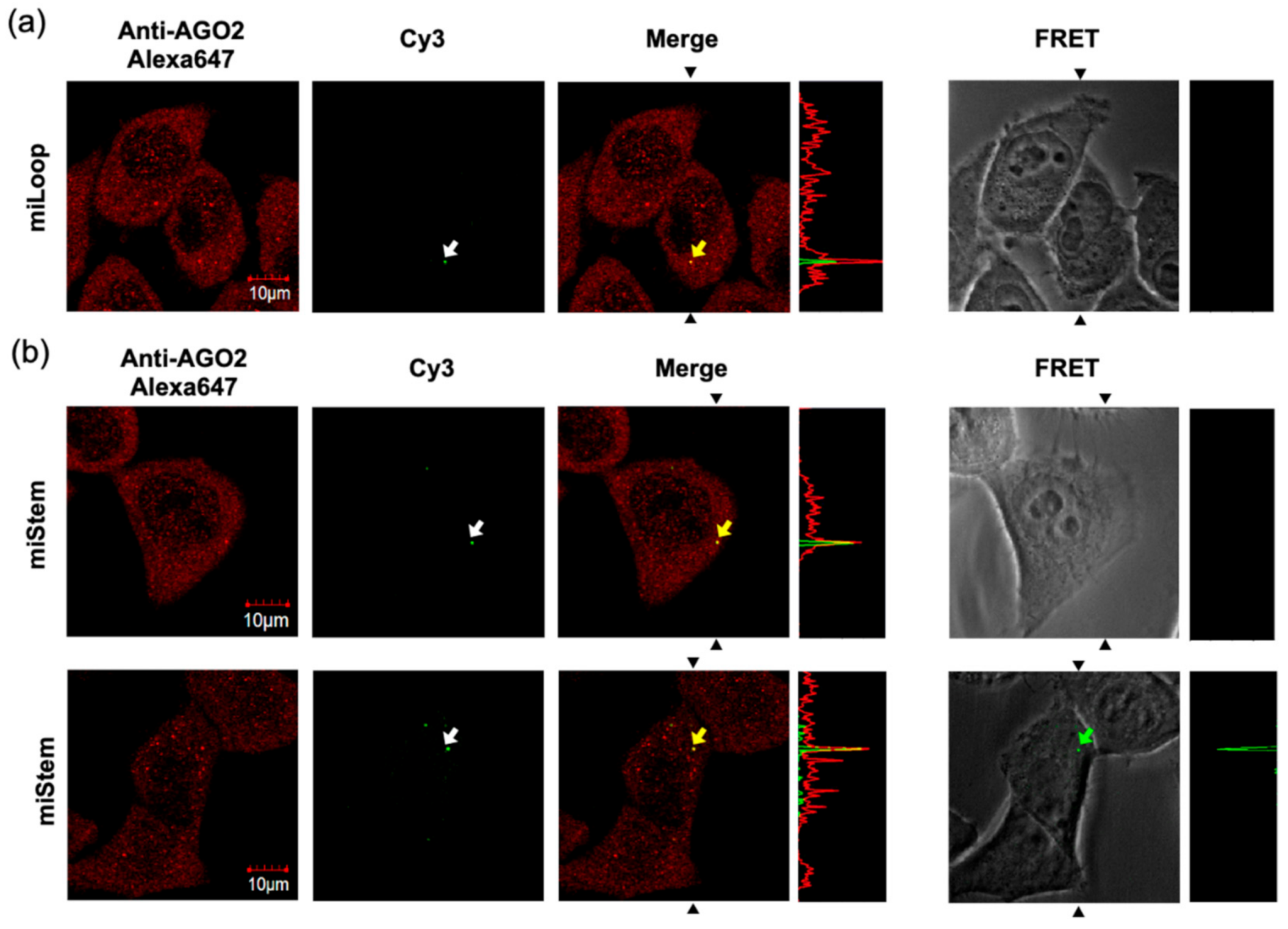
3.4. Cell Imaging Analyses of Fluorescently Modified Pre-miR17s
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwakawa, H.O.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Kasinski, A.L.; Slack, F.J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 2014, 24, R762–R776. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef]
- Baumann, V.; Winkler, J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014, 6, 1967–1984. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Watts, J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017, 35, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Leung, A.K.L. The Whereabouts of microRNA Actions: Cytoplasm and Beyond. Trends Cell Biol. 2015, 25, 601–610. [Google Scholar] [CrossRef]
- Sen, G.L.; Blau, H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005, 7, 633–636. [Google Scholar] [CrossRef]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tsourkas, A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009, 37, e100. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.B.; Bao, G.; Searles, C.D. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012, 40, e13. [Google Scholar] [CrossRef]
- Bidar, N.; Oroojalian, F.; Baradaran, B.; Eyvazi, S.; Amini, M.; Jebelli, A.; Hosseini, S.S.; Pashazadeh-Panahi, P.; Mokhtarzadeh, A.; de la Guardia, M. Monitoring of microRNA using molecular beacons approaches: Recent advances. Trac-Trend. Anal. Chem. 2020, 131, 116021. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Ali, A.; Chu, C.Y.; Cao, H.; Rana, T.M. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem. Biol. 2004, 11, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stoter, M.; et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef]
- Wittrup, A.; Ai, A.; Liu, X.; Hamar, P.; Trifonova, R.; Charisse, K.; Manoharan, M.; Kirchhausen, T.; Lieberman, J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015, 33, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Pitchiaya, S.; Androsavich, J.R.; Walter, N.G. Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep. 2012, 13, 709–715. [Google Scholar] [CrossRef]
- Holzhauser, C.; Liebl, R.; Goepferich, A.; Wagenknecht, H.A.; Breunig, M. RNA “traffic lights”: An analytical tool to monitor siRNA integrity. ACS Chem. Biol. 2013, 8, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, M.; Helm, M. Live cell imaging of duplex siRNA intracellular trafficking. Nucleic Acids Res. 2015, 43, 4650–4660. [Google Scholar] [CrossRef] [PubMed]
- Pitchiaya, S.; Heinicke, L.A.; Park, J.I.; Cameron, E.L.; Walter, N.G. Resolving Subcellular miRNA Trafficking and Turnover at Single-Molecule Resolution. Cell Rep. 2017, 19, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, Y.; Ito, A.; Ito, H.; Urushihara, M.; Takai, J.; Fujii, T.; Liang, X.G.; Kashida, H.; Asanuma, H. Selective labeling of mature RISC using a siRNA carrying fluorophore-quencher pair. Chem. Sci. 2013, 4, 4016–4021. [Google Scholar] [CrossRef]
- Li, S.B.; Ma, D.J.; Yi, L.; Mei, S.Y.; Ouyang, D.; Xi, Z. Terminal dual-labeling of a transcribed RNA. Bioorg. Med. Chem. Lett. 2013, 23, 6304–6306. [Google Scholar] [CrossRef]
- Astakhova, I.K.; Wengel, J. Interfacing Click Chemistry with Automated Oligonucleotide Synthesis for the Preparation of Fluorescent DNA Probes Containing Internal Xanthene and Cyanine Dyes. Chem. Eur. J. 2013, 19, 1112–1122. [Google Scholar] [CrossRef]
- Pradere, U.; Brunschweiger, A.; Gebert, L.F.R.; Lucic, M.; Roos, M.; Hall, J. Chemical Synthesis of Mono- and Bis-Labeled Pre-MicroRNAs. Angew. Chem. Int. Ed. 2013, 52, 12028–12032. [Google Scholar] [CrossRef]
- Pradere, U.; Hall, J. Site-Specific Difunctionalization of Structured RNAs Yields Probes for microRNA Maturation. Bioconjug. Chem. 2016, 27, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Kashida, H.; Kamiya, Y. De Novo Design of Functional Oligonucleotides with Acyclic Scaffolds. Chem. Rec. 2014, 14, 1055–1069. [Google Scholar] [CrossRef]
- Asanuma, H.; Murayama, K.; Kamiya, Y.; Kashida, H. Design of photofunctional oligonucleotides by copolymerization of natural nucleobases with base surrogates prepared from acyclic scaffolds. Polym. J. 2017, 49, 279–289. [Google Scholar] [CrossRef]
- Asanuma, H.; Akahane, M.; Kondo, N.; Osawa, T.; Kato, T.; Kashida, H. Quencher-free linear probe with multiple fluorophores on an acyclic scaffold. Chem. Sci. 2012, 3, 3165–3169. [Google Scholar] [CrossRef]
- Fujii, T.; Urushihara, M.; Kashida, H.; Ito, H.; Liang, X.G.; Yagi-Utsumi, M.; Kato, K.; Asanuma, H. Reversed Assembly of Dyes in an RNA Duplex Compared with Those in DNA. Chem. Euro. J. 2012, 18, 13304–13313. [Google Scholar] [CrossRef] [PubMed]
- Murayama, K.; Kamiya, Y.; Kashida, H.; Asanuma, H. Ultrasensitive Molecular Beacon Designed with Totally Serinol Nucleic Acid (SNA) for Monitoring mRNA in Cells. ChemBioChem 2015, 16, 1298–1301. [Google Scholar] [CrossRef]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Upton, J.P.; Wang, L.; Han, D.; Wang, E.S.; Huskey, N.E.; Lim, L.; Truitt, M.; McManus, M.T.; Ruggero, D.; Goga, A.; et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 2012, 338, 818–822. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamiya, Y.; Kamimoto, H.; Zhu, H.; Asanuma, H. Development and Modification of Pre-miRNAs with a FRET Dye Pair for the Intracellular Visualization of Processing Intermediates That Are Generated in Cells. Sensors 2021, 21, 1785. https://doi.org/10.3390/s21051785
Kamiya Y, Kamimoto H, Zhu H, Asanuma H. Development and Modification of Pre-miRNAs with a FRET Dye Pair for the Intracellular Visualization of Processing Intermediates That Are Generated in Cells. Sensors. 2021; 21(5):1785. https://doi.org/10.3390/s21051785
Chicago/Turabian StyleKamiya, Yukiko, Hiroshi Kamimoto, Hongyu Zhu, and Hiroyuki Asanuma. 2021. "Development and Modification of Pre-miRNAs with a FRET Dye Pair for the Intracellular Visualization of Processing Intermediates That Are Generated in Cells" Sensors 21, no. 5: 1785. https://doi.org/10.3390/s21051785
APA StyleKamiya, Y., Kamimoto, H., Zhu, H., & Asanuma, H. (2021). Development and Modification of Pre-miRNAs with a FRET Dye Pair for the Intracellular Visualization of Processing Intermediates That Are Generated in Cells. Sensors, 21(5), 1785. https://doi.org/10.3390/s21051785





