Sepsis Mortality Prediction Using Wearable Monitoring in Low–Middle Income Countries
Abstract
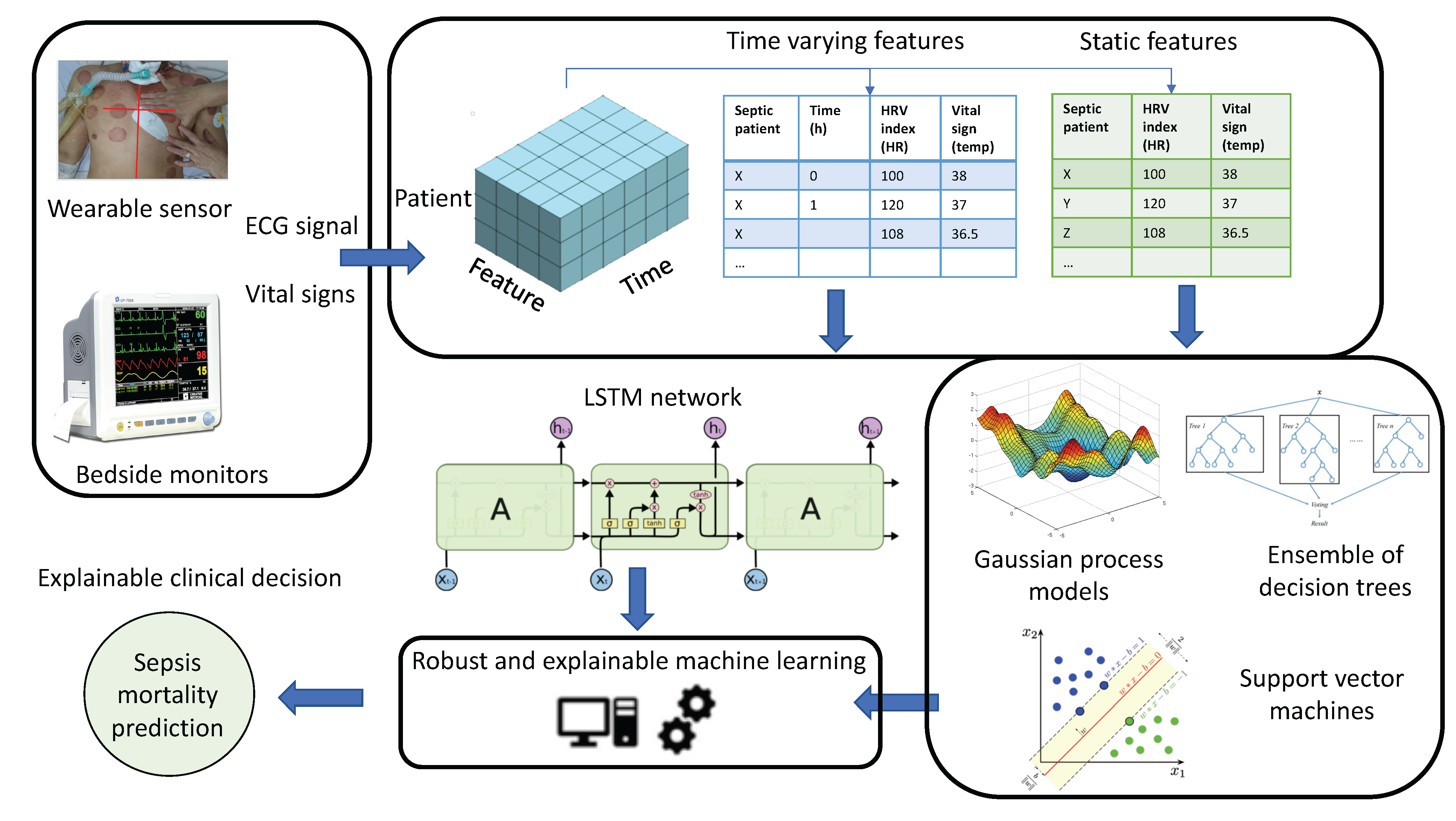
:1. Introduction
- We gathered a rich dataset of simultaneous long-term ECG recordings from wearable sensors and vital signs from bedside monitoring systems from adult patients with sepsis admitted to the hospital within a resource-limited healthcare system.
- We demonstrate the high potential of physiological data gathered from wearable devices compared to traditional bedside monitoring systems for sepsis management.
- We propose an interpretable automatic ML-based solution for a long-term prediction of hospital discharge outcome in critically ill sepsis patients using the data collected during the first day of admission to be potentially implemented in practice.
2. Related Works
3. Materials and Methods
3.1. Study Participants
3.2. Experimental Protocol (Data Collection)
3.3. Final Cohort of Patients
3.4. Processing Pipeline
3.4.1. Data Collection and Feature Extraction
3.4.2. Machine Learning Training
- = [HR(mean), HR(std),RR(mean), RR(std), RMSSD, , , , , , , SD1, SD2, SD1/SD2, SampEn, , ]
- = [Temp, Pulse, SBP, Resp, SP02]
- = [,]
- Support vector machines with recursive feature elimination (SVM-RFE): SVM models are powerful classification tools aiming to find a hyperplane that maximizes the distance between binary labelled observation samples [41]. We applied the standard nonlinear SVM with the radial basis function (RBF) kernel embedded with recursive feature elimination (RFE) on our dataset. We give further details on the embedded RFE algorithm for feature selection in the next subsection. We used the Libsvm package to apply the SVM-RFE models [42].
- Gaussian process classification model: Gaussian process classification (GPC) models are a class of machine learning models which are based on non-parametric Bayesian formulation. In GPC settings, a latent variable that represents the classification logit is defined and a prior distribution is placed over the latent space in the form of a Gaussian process (GP) [43,44]. We used the Gaussian Processes for Machine Learning (GPML) toolbox to implement the GPC model training in this study [45]. We chose a linear mean function as the prior function of GP model and used the square exponential function for the covariance function.
- Gradient Boosting Decision Tree: Gradient boosting decision tree (GBDT) is an ensemble model of decision trees in which each decision tree is sequentially built on the gradient descent direction of a loss function. In each iteration, GBDT learns the decision trees by fitting the negative gradients known as residual errors [46].In this paper, we used the software library, eXtreme Gradient Boosting (XGBoost), which is an implementation of GBDT in Python designed for speed and performance [47]. Tuning the XGBoost can be a very daunting task because of the number of hyperparameters it has. We applied grid search with reasonable ranges on only two of the parameters, the number of trees and the maximum tree depth. All the possible combinations of these two parameter values are run for the model tuning and the one with best performance is retained as the optimal values. The rest of the parameters were kept as default in XGBoost library. The final values for the number of trees and the maximum tree depth were set to 4 and 3, respectively.
3.4.3. Machine Learning Interpretation
- RFE for SVM classifier: RFE is an embedded feature selection method based on a backward sequential selection that eliminates a feature in a feature set of size m that has the least effect on the SVM weight-vector norm at each iteration [50]. This way, the features are ranked and the SVM classification is repeated m times while the last ranked features are removed. Finally, a subset of features with size r that optimises the performance of the SVM classifier are selected.
- GP interpretability framework: We applied a recently developed interpretability analysis of GPC models, based on an explicit form of the GP inference equations to quantify the importance of each feature contributing to the GPC model prediction [51]. Within this framework, small perturbations are propagated to each data input in succession through the prior model and then the GP posterior, in order to quantify the contribution of each feature input to the overall model prediction of a data sample. In particular, given a GP model trained on a given dataset, a test input point and a neighbourhood around the latter, we compute the probability that there exists a point in the neighbourhood such that the prediction of the GP on the latter differs from the initial test input point by at least a given threshold. The outcome is an interpretability metric, denoted , that corresponds to the importance of each data point in the model training.Let us define as a generic input data sample in our dataset where is the sub-vector of x that includes only the indices of and L is the total number of features. For any test sample data with a subset of indices l, a norm , and a radius , we perform a set of perturbations of magnitude up to around according to Equation (2).Then, we define the interpretability metric according to Equation (3) which reflects how much local perturbations of the indices l of can change the prediction probabilities.where encodes the probability that x belongs to class 1. Detailed mathematical formulation of this framework can be found in [51,52].
- Feature importance in GBDT: Decision trees bring the benefit of interpretability by means of decision analysis on the structure of the trees. One of the main features of GBDT algorithms is that they identify attributes that contribute the most towards the performance. We quantified the importance of each feature based on the number of times a feature is used to split the data across all trees.Within XGBoost library, a feature importance score can be obtained based on the relative contribution of the corresponding feature to the model calculated by taking each feature’s contribution for each tree in the model [46]. A higher value of this metric when compared to another feature implies it is more important for generating a prediction. We selected the “gain” value to report the feature importance results.
- Interpretable model-agnostic explanation for LSTM: In recent years, among the deep-learning methods, local interpretable model-agnostic explanations (LIME) [53] has emerged as a new evaluation method that can explain the predictions of any classifier by approximating it locally with an interpretable model. It builds a local linear approximation of a complex model’s behaviour in the neighbourhood of a data sample by treating the model as a black box and classifying near permutations of the data sample being explained. Therefore, the output of LIME is a list of explanations, reflecting the contribution of each feature to the prediction of a data sample. This provides local interpretability and allows the determination of the features with the most important impact on the prediction of the data sample.For tabular data, variations of the data are produced by perturbing each feature individually. In particular, we applied the TabularLIME algorithm from the LIME package in Python to quantify the importance of each feature at each timestamp for the trained LSTM model.
4. Results
4.1. Results with Static Features
- Precision (PPV): The percentage of truly positive predictions out of the positive predicted.
- F1-score: Harmonic mean of precision and recall where recall is the percentage of predicted positive out of the total positive. This metric takes both false positive and false negatives into account.
- AUCROC: The area under the curve of receiver operating characteristic curve.
- AUCPRC: The area under the curve of precision recall curve.
4.2. Results with Time-Varying Features
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Fleischmann, C.; Scherag, A.; Adhikari, N.K.; Hartog, C.S.; Tsaganos, T.; Schlattmann, P.; Angus, D.C.; Reinhart, K. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.R.; Angus, D.C. Trying to improve sepsis care in low-resource settings. JAMA 2017, 318, 1225–1227. [Google Scholar] [CrossRef]
- Olufadewa, I.; Adesina, M.; Ayorinde, T. Global health in low-income and middle-income countries: A framework for action. Lancet Glob. Health 2021, 9, e899–e900. [Google Scholar] [CrossRef]
- Rello, J.; Leblebicioglu, H. Sepsis and septic shock in low-income and middle-income countries: Need for a different paradigm. Int. J. Infect. Dis. 2016, 48, 120–122. [Google Scholar] [CrossRef] [Green Version]
- Dat, V.Q.; Long, N.T.; Giang, K.B.; Diep, P.B.; Giang, T.H.; Diaz, J.V. Healthcare infrastructure capacity to respond to severe acute respiratory infection (SARI) and sepsis in Vietnam: A low-middle income country. J. Crit. Care 2017, 42, 109–115. [Google Scholar] [CrossRef]
- Kiyasseh, D.; Zhu, T.; Clifton, D. The Promise of Clinical Decision Support Systems Targetting Low-Resource Settings. IEEE Rev. Biomed. Eng. 2020, 15, 354–371. [Google Scholar] [CrossRef]
- Kim, M.; Ahn, S.; Kim, W.Y.; Sohn, C.H.; Seo, D.W.; Lee, Y.S.; Lim, K.S. Predictive performance of the quick Sequential Organ Failure Assessment score as a screening tool for sepsis, mortality, and intensive care unit admission in patients with febrile neutropenia. Support. Care Cancer 2017, 25, 1557–1562. [Google Scholar] [CrossRef]
- Lim, W.T.; Fang, A.H.; Loo, C.M.; Wong, K.S.; Balakrishnan, T. Use of the National Early Warning Score (NEWS) to identify acutely deteriorating patients with sepsis in acute medical ward. Ann. Acad. Med. Singap. 2019, 48, 145–149. [Google Scholar]
- Wang, R.; Blackburn, G.; Desai, M.; Phelan, D.; Gillinov, L.; Houghtaling, P.; Gillinov, M. Accuracy of wrist-worn heart rate monitors. JAMA Cardiol. 2017, 2, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Ming, D.K.; Sangkaew, S.; Chanh, H.Q.; Nhat, P.T.; Yacoub, S.; Georgiou, P.; Holmes, A.H. Continuous physiological monitoring using wearable technology to inform individual management of infectious diseases, public health and outbreak responses. Int. J. Infect. Dis. 2020, 96, 648–654. [Google Scholar] [CrossRef]
- Joshi, M.; Ashrafian, H.; Aufegger, L.; Khan, S.; Arora, S.; Cooke, G.; Darzi, A. Wearable sensors to improve detection of patient deterioration. Expert Rev. Med. Devices 2019, 16, 145–154. [Google Scholar] [CrossRef]
- Breteler, M.J.; KleinJan, E.J.; Dohmen, D.A.; Leenen, L.P.; van Hillegersberg, R.; Ruurda, J.P.; van Loon, K.; Blokhuis, T.J.; Kalkman, C.J. Vital signs monitoring with wearable sensors in high-risk surgical patients: A clinical validation study. Anesthesiology 2020, 132, 424–439. [Google Scholar] [CrossRef]
- Downey, C.; Randell, R.; Brown, J.; Jayne, D.G. Continuous versus intermittent vital signs monitoring using a wearable, wireless patch in patients admitted to surgical wards: Pilot cluster randomized controlled trial. J. Med. Internet Res. 2018, 20, e10802. [Google Scholar] [CrossRef]
- Quinten, V.M.; van Meurs, M.; Renes, M.H.; Ligtenberg, J.J.; Ter Maaten, J.C. Protocol of the sepsivit study: A prospective observational study to determine whether continuous heart rate variability measurement during the first 48 h of hospitalisation provides an early warning for deterioration in patients presenting with infection or sepsis to the emergency department of a Dutch academic teaching hospital. BMJ Open 2017, 7, e018259. [Google Scholar]
- Edgcombe, H.; Paton, C.; English, M. Enhancing emergency care in low-income countries using mobile technology-based training tools. Arch. Dis. Child. 2016, 101, 1149–1152. [Google Scholar] [CrossRef] [Green Version]
- Steinhubl, S.R.; Feye, D.; Levine, A.C.; Conkright, C.; Wegerich, S.W.; Conkright, G. Validation of a portable, deployable system for continuous vital sign monitoring using a multiparametric wearable sensor and personalised analytics in an Ebola treatment centre. BMJ Glob. Health 2016, 1, e000070. [Google Scholar] [CrossRef] [Green Version]
- Garbern, S.C.; Mbanjumucyo, G.; Umuhoza, C.; Sharma, V.K.; Mackey, J.; Tang, O.; Martin, K.D.; Twagirumukiza, F.R.; Rosman, S.L.; McCall, N.; et al. Validation of a wearable biosensor device for vital sign monitoring in septic emergency department patients in Rwanda. Digit. Health 2019, 5, 2055207619879349. [Google Scholar] [CrossRef]
- de Castilho, F.M.; Ribeiro, A.L.P.; Nobre, V.; Barros, G.; de Sousa, M.R. Heart rate variability as predictor of mortality in sepsis: A systematic review. PLoS ONE 2018, 13, e0203487. [Google Scholar]
- de Castilho, F.M.; Ribeiro, A.L.P.; da Silva, J.L.P.; Nobre, V.; de Sousa, M.R. Heart rate variability as predictor of mortality in sepsis: A prospective cohort study. PLoS ONE 2017, 12, e0180060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Doorn, W.P.; Stassen, P.M.; Borggreve, H.F.; Schalkwijk, M.J.; Stoffers, J.; Bekers, O.; Meex, S.J. A comparison of machine learning models versus clinical evaluation for mortality prediction in patients with sepsis. PLoS ONE 2021, 16, e0245157. [Google Scholar] [CrossRef] [PubMed]
- Chiew, C.J.; Liu, N.; Tagami, T.; Wong, T.H.; Koh, Z.X.; Ong, M.E. Heart rate variability based machine learning models for risk prediction of suspected sepsis patients in the emergency department. Medicine 2019, 98, e14197. [Google Scholar] [CrossRef] [PubMed]
- Burykin, A.; Peck, T.; Krejci, V.; Vannucci, A.; Kangrga, I.; Buchman, T.G. Toward optimal display of physiologic status in critical care: I. Recreating bedside displays from archived physiologic data. J. Crit. Care 2011, 26, 105.e1–105.e9. [Google Scholar] [CrossRef]
- Gircys, R.; Kazanavicius, E.; Maskeliunas, R.; Damasevicius, R.; Wozniak, M. Wearable system for real-time monitoring of hemodynamic parameters: Implementation and evaluation. Biomed. Signal Process. Control. 2020, 59, 101873. [Google Scholar] [CrossRef]
- Odusami, M.; Misra, S.; Abayomi-Alli, O.; Olamilekan, S.; Moses, C. An Enhanced IoT-Based Array of Sensors for Monitoring Patients’ Health. In Intelligent Internet of Things for Healthcare and Industry; Springer: Berlin, Germany, 2022; pp. 105–125. [Google Scholar]
- Van, H.M.T.; Van Hao, N.; Quoc, K.P.N.; Hai, H.B.; Yen, L.M.; Nhat, P.T.H.; Duong, H.T.H.; Thuy, D.B.; Zhu, T.; Greeff, H.; et al. Vital sign monitoring using wearable devices in a Vietnamese intensive care unit. BMJ Innov. 2021, 7, 7–11. [Google Scholar] [CrossRef]
- Taylor, R.A.; Pare, J.R.; Venkatesh, A.K.; Mowafi, H.; Melnick, E.R.; Fleischman, W.; Hall, M.K. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big data–driven, machine learning approach. Acad. Emerg. Med. 2016, 23, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Vorwerk, C.; Loryman, B.; Coats, T.; Stephenson, J.; Gray, L.; Reddy, G.; Florence, L.; Butler, N. Prediction of mortality in adult emergency department patients with sepsis. Emerg. Med. J. 2009, 26, 254–258. [Google Scholar] [CrossRef]
- Perng, J.W.; Kao, I.H.; Kung, C.T.; Hung, S.C.; Lai, Y.H.; Su, C.M. Mortality prediction of septic patients in the emergency department based on machine learning. J. Clin. Med. 2019, 8, 1906. [Google Scholar] [CrossRef] [Green Version]
- Barnaby, D.P.; Fernando, S.M.; Herry, C.L.; Scales, N.B.; Gallagher, E.J.; Seely, A.J. Heart rate variability, clinical and laboratory measures to predict future deterioration in patients presenting with sepsis. Shock 2019, 51, 416–422. [Google Scholar] [CrossRef]
- Cedillo, J.L.; Arnalich, F.; Martín-Sánchez, C.; Quesada, A.; Rios, J.J.; Maldifassi, M.C.; Atienza, G.; Renart, J.; Fernández-Capitán, C.; García-Rio, F.; et al. Usefulness of α7 nicotinic receptor messenger RNA levels in peripheral blood mononuclear cells as a marker for cholinergic antiinflammatory pathway activity in septic patients: Results of a pilot study. J. Infect. Dis. 2015, 211, 146–155. [Google Scholar] [CrossRef] [Green Version]
- Nogueira, A.C.; Kawabata, V.; Biselli, P.; Lins, M.H.; Valeri, C.; Seckler, M.; Hoshino, W.; Júnior, L.G.; Bernik, M.M.S.; de Andrade Machado, J.B.; et al. Changes in plasma free fatty acid levels in septic patients are associated with cardiac damage and reduction in heart rate variability. Shock 2008, 29, 342–348. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.L.; Shen, Y.S.; Huang, C.C.; Chen, J.H.; Kuo, C.D. Postresuscitation autonomic nervous modulation after cardiac arrest resembles that of severe sepsis. Am. J. Emerg. Med. 2012, 30, 143–150. [Google Scholar] [CrossRef]
- Duque, M.G.; Olivera, C.E.; Torres, E.P.; Durán, O.S.; Estrada, V.N. ECAIS study: Inadvertent cardiovascular adverse events in sepsis. Med. Intensiv. 2012, 36, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Tateishi, Y.; Oda, S.; Nakamura, M.; Watanabe, K.; Kuwaki, T.; Moriguchi, T.; Hirasawa, H. Depressed heart rate variability is associated with high IL-6 blood level and decline in the blood pressure in septic patients. Shock 2007, 28, 549–553. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Gomes, P.; Margaritoff, P.; Silva, H. pyHRV: Development and evaluation of an open-source python toolbox for heart rate variability (HRV). In Proceedings of the International Conference on Electrical, Electronic and Computing Engineering (ICETRAN), Silver Lake, Serbia, 3–6 June 2019; pp. 822–828. [Google Scholar]
- Shu, L.; Xie, J.; Yang, M.; Li, Z.; Li, Z.; Liao, D.; Xu, X.; Yang, X. A review of emotion recognition using physiological signals. Sensors 2018, 18, 2074. [Google Scholar] [CrossRef] [Green Version]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. Acm Trans. Intell. Syst. Technol. (Tist) 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Rasmussen, C.E. Gaussian processes in machine learning. In Summer School on Machine Learning; Springer: Berlin, Germany, 2003; pp. 63–71. [Google Scholar]
- Ghiasi, S.; Patane, A.; Greco, A.; Laurenti, L.; Scilingo, E.P.; Kwiatkowska, M. Gaussian Processes with Physiologically-Inspired Priors for Physical Arousal Recognition. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 54–57. [Google Scholar]
- Rasmussen, C.E.; Nickisch, H. Gaussian processes for machine learning (GPML) toolbox. J. Mach. Learn. Res. 2010, 11, 3011–3015. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Scm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Brownlee, J. XGBoost with Python: Gradient Boosted Trees with XGBoost and Scikit-Learn; Machine Learning Mastery: San Juan, PR, USA, 2016. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Ketkar, N. Introduction to feature selection. In Deep Learning with Python; Springer: New York, NY, USA, 2017; pp. 97–111. [Google Scholar]
- Weston, J.; Mukherjee, S.; Chapelle, O.; Pontil, M.; Poggio, T.; Vapnik, V. Feature selection for SVMs. In Proceedings of the Advances in Neural Information Processing Systems (NIPS), Denver, CO, USA, 29 November–4 December 1999; Volume 13. [Google Scholar]
- Cardelli, L.; Kwiatkowska, M.; Laurenti, L.; Patane, A. Robustness guarantees for Bayesian inference with Gaussian processes. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 7759–7768. [Google Scholar]
- Ghiasi, S.; Patane, A.; Greco, A.; Laurenti, L.; Gentili, C.; Scilingo, E.P.; Kwiatkowska, M. Physiologically-informed gaussian processes for interpretable modelling of psycho-physiological states. TechRxiv 2022. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]




| Variable | All (n = 40) | Death (n = 14) | Survival (n = 26) |
|---|---|---|---|
| gender (M) | 67.5 % | 64.3 % | 72 % |
| Age (>=64) | n = 12 | n = 2 | n = 10 |
| Age (50–64) | n = 7 | n = 1 | n = 6 |
| Age (<50) | n = 21 | n = 11 | n = 10 |
| Hospital length of stay | 12.08 ± 12.25 | 10.14 ± 15.97 | 13.04 ± 9.9 |
| SOFA at admission | 2.13 ± 1.74 | 1.77 ± 1.58 | 2.78 ± 1.89 |
| Parameter | Unit | Description |
|---|---|---|
| HRV time parameters | ||
| HR(mean) | BPM | Mean of heart rate |
| HR(std) | BPM | Standard deviation of heart rate |
| RR(mean) | ms | Mean of RR intervals |
| RR(std) | ms | Standard deviation of RR intervals |
| RMSSD | ms | Root mean square of successive |
| RR interval differences | ||
| HRV frequency parameters | ||
| ms2 | Absolute power in VLF band | |
| ms2 | Absolute power in LF band | |
| ms2 | Absolute power in HF band | |
| Hz | Frequency where maximum power | |
| occurs in VLF band | ||
| Hz | Frequency where maximum power | |
| occurs in LF band | ||
| Hz | Frequency where maximum power | |
| occurs in HF band | ||
| HRV nonlinear parameters | ||
| SD1 | ms | Standard deviation along the minor axis |
| in Poincare plot | ||
| SD2 | ms | Standard deviation along the major axis |
| in Poincare plot | ||
| SD1/SD2 | - | Ratio between SD1 & SD2 |
| S | - | Area of the fitted ellipse (Poincare plot) |
| SampEn | - | Sample entropy of RR series |
| - | Alpha value of the short term fluctuations | |
| in detrended fluctuation analysis | ||
| - | Alpha value of the long term fluctuations | |
| in detrended fluctuation analysis | ||
| Vital signs | ||
| Temp | °C | Temperature |
| Pulse | BPM | Hear rate |
| SBP | mmHG | Systolic blood pressure |
| Resp | BPM | Respiratory rate |
| SP02 | % | Peripheral capillary oxygen saturation |
| SVM-RFE | Gaussian Process | XGBoost | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PPV (LOSO) | |||||||||
| PPV (SKfold) | |||||||||
| F1-score (LOSO) | |||||||||
| F1-score (SKfold) | |||||||||
| AUCROC (LOSO) | |||||||||
| AUCROC (SKfold) | |||||||||
| AUCPRC (LOSO) | |||||||||
| AUCPRC (SKfold) | |||||||||
| AUCROC | 0.70 | 0.62 | 0.67 | 0.68 |
| AUCPRC | 0.83 | 0.72 | 0.81 | 0.82 |
| SVM | |||
| Gaussian Process | |||
| XGBoost |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghiasi, S.; Zhu, T.; Lu, P.; Hagenah, J.; Khanh, P.N.Q.; Hao, N.V.; Vital Consortium; Thwaites, L.; Clifton, D.A. Sepsis Mortality Prediction Using Wearable Monitoring in Low–Middle Income Countries. Sensors 2022, 22, 3866. https://doi.org/10.3390/s22103866
Ghiasi S, Zhu T, Lu P, Hagenah J, Khanh PNQ, Hao NV, Vital Consortium, Thwaites L, Clifton DA. Sepsis Mortality Prediction Using Wearable Monitoring in Low–Middle Income Countries. Sensors. 2022; 22(10):3866. https://doi.org/10.3390/s22103866
Chicago/Turabian StyleGhiasi, Shadi, Tingting Zhu, Ping Lu, Jannis Hagenah, Phan Nguyen Quoc Khanh, Nguyen Van Hao, Vital Consortium, Louise Thwaites, and David A. Clifton. 2022. "Sepsis Mortality Prediction Using Wearable Monitoring in Low–Middle Income Countries" Sensors 22, no. 10: 3866. https://doi.org/10.3390/s22103866






