Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders
Abstract
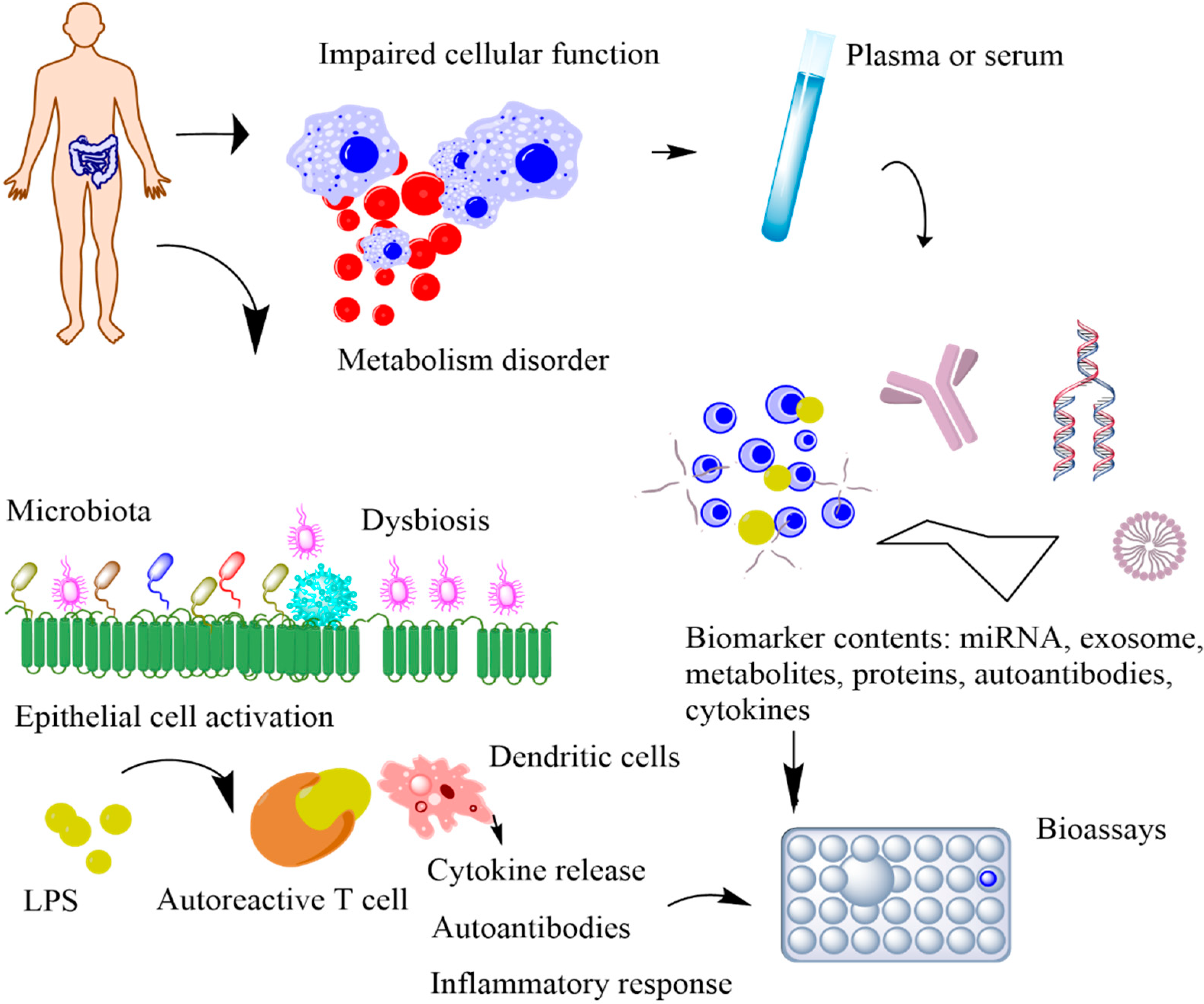
1. Introduction
2. Biomarkers for Metabolic Syndrome
2.1. Metabolic Biomarkers for Predicting Cardiovascular Disease
2.2. Metabolic Biomarkers for Prediabetes
2.3. Metabolic Biomarkers for Cancer
| Metabolic Syndrome | Biomarker | Clinical Approval | Concentration | Refs. |
|---|---|---|---|---|
| Cardiovascular Diseases (CVDs) | C-reactive Protein (CRP) | Approved | >3 mg L−1 | [37] |
| Highly sensitive CRP | <1 mg L−1 | [38] | ||
| Cardiac troponin 1 (cTn1) | Approved | >0.5 μg L−1 | [39] | |
| Procalcitonin | >67.89 μg L−1 | |||
| Cholesterol | Approved | >240 mg dL−1 | [40] | |
| LDL cholesterol | Approved | >130 mg dL−1 | [41] | |
| HDL cholesterol | Approved | <40 mg dL−1 | [42] | |
| Triglyceride | >150 mg dL−1 | [43] | ||
| Diabetes | Glucose | Approved | >125 mg dL−1 | [44] |
| CD14 | [45] | |||
| CD99 | [46] | |||
| HbA1c | Approved | >6.5% | [24] | |
| GA | >16.9% | [47] | ||
| Adiponectin | <6 mg mL−1 | [48] | ||
| Fructosamine | Approved | <2.5 mmol L−1 | [24] | |
| Cancer | Fumarate | Approved | >1.35 mcg mg−1 creatinine | [49] |
| 2-hydroxyglutarate | Approved | >700 ng mL−1 | [50] | |
| Sarcosine | Approved | >5000 nM | [51] | |
| Polyamines | Approved | 35 kU L−1 | [52] | |
| Lactate | Approved | >1.8 mmol L−1 | [53] | |
| Lactate dehydrogenase | >280 U L−1 | [54] | ||
| Autoimmune disease | Hydrogen peroxide (H2O2), hydroxyl radical (OH), superoxide anion radical (O2−), and nitric oxide (NO) | (investigating) | 308 ppb (cutoff of 77 nL mL−1) | [55] |
| Serum fatty acids (monounsaturated fatty acids such as lauric acid (C12:0), myristic acid (C14:0), stearic acid (C18:0), lignoceric acid (C24:0), palmitic acid (C16:0) and heptadecanoic acid (C17:0); saturated fatty acids, Cis-10-pentadecanoic acid (C15:1), Cis-11-eicosenoic acid (C20:1n9), and erucic acid (C22:1n9) as well as the gamma-linolenic acid (C18:3n6) polyunsaturated fatty acid)) | (investigating) | 86.7% specificity (ROC analysis) | [56] | |
| Serum fatty acid (3-hydroxypropionic and methylcitric acids, propionylglycine, tiglylglycine, 3-hydroxy-n-valeric, and 3-keto-n-valeric acids) | investigating | 0.856 (ROC analysis) | [56] |
2.4. Metabolic Biomarkers for Autoimmune Disease
3. Non-Invasively Accessible Resources for Biomarkers
3.1. Sweat
3.2. Tears
3.3. Breath
3.4. Saliva
3.5. Urine
4. Biosensor Platforms
4.1. Electrochemical Biosensors
4.1.1. Cardiovascular Disease
4.1.2. Prediabetes
4.1.3. Cancer
4.1.4. Autoimmune Disease
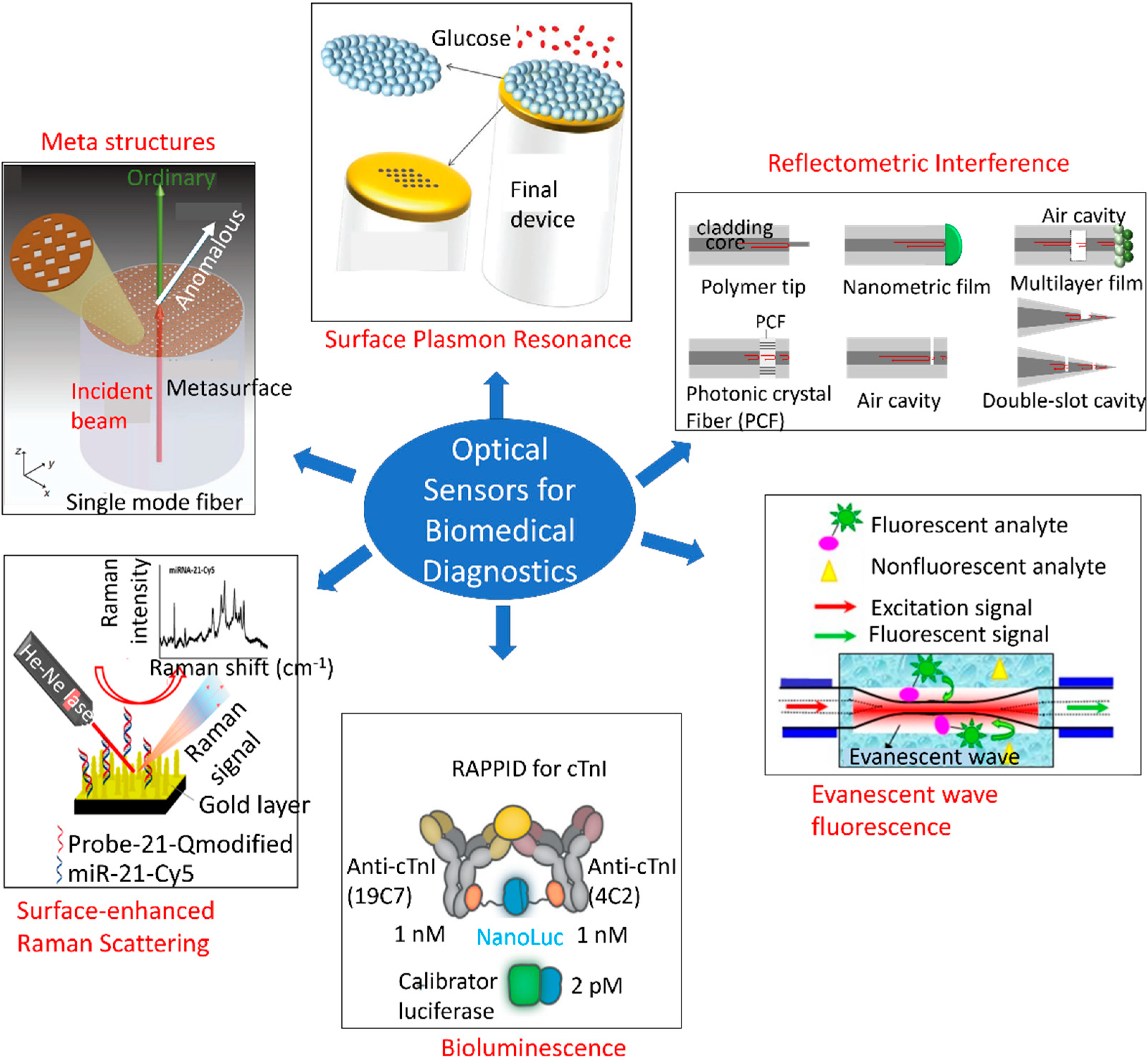
4.2. Optical Biosensors
4.2.1. Multiplexed Optical Detection Systems
4.2.2. Cardiovascular Disease
4.2.3. Prediabetes
4.2.4. Cancer
4.2.5. Autoimmune Disease
5. Outlook: Towards Multimodal Sensor Platforms
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lima, L.F.; Braga, V.D.A.; Silva, M.E.S.D.F.; Cruz, J.D.C.; Santos, S.H.S.; Monteiro, M.M.D.O.; Balarini, C.D.M. Adipokines, diabetes and atherosclerosis: An inflammatory association. Front. Physiol. 2015, 6, 304. [Google Scholar] [CrossRef]
- Mendrick, D.L.; Diehl, A.M.; Topor, L.S.; Dietert, R.R.; Will, Y.; La Merrill, A.M.; Bouret, S.; Varma, V.; Hastings, K.L.; Schug, T.T.; et al. Metabolic Syndrome and Associated Diseases: From the Bench to the Clinic. Toxicol. Sci. 2018, 162, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Srikanthan, K.; Feyh, A.; Visweshwar, H.; Shapiro, J.I.; Sodhi, K. Systematic Review of Metabolic Syndrome Biomarkers: A Panel for Early Detection, Management, and Risk Stratification in the West Virginian Population. Int. J. Med. Sci. 2016, 13, 25–38. [Google Scholar] [CrossRef]
- Stefan, N.; Birkenfeld, A.L.; Schulze, M.B. Global pandemics interconnected–obesity, impaired metabolic health and COVID-19. Nat. Rev. Endocrinol. 2021, 17, 135–149. [Google Scholar] [CrossRef]
- Han, T.S.; Lean, M.E. A clinical perspective of obesity, metabolic syndrome and cardiovascular disease. JRSM Cardiovasc. Dis. 2016, 5, 2048004016633371. [Google Scholar] [CrossRef]
- Sasya, M.; Devi, K.S.S.; Babu, J.K.; Rayappan, J.B.B.; Krishnan, U.M. Metabolic Syndrome—An Emerging Constellation of Risk Factors: Electrochemical Detection Strategies. Sensors 2020, 20, 103. [Google Scholar] [CrossRef]
- Ates, H.C.; Yetisen, A.K.; Güder, F.; Dincer, C. Wearable devices for the detection of COVID-19. Nat. Electron. 2021, 4, 13–14. [Google Scholar] [CrossRef]
- Sia, S.K.; Kricka, L.J. Microfluidics and point-of-care testing. Lab Chip 2008, 8, 1982–1983. [Google Scholar] [CrossRef]
- Jain, K.K. Biomarkers of Infectious Diseases. In The Handbook of Biomarkers; Springer: Berlin/Heidelberg, Germany, 2017; pp. 219–238. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Escrig, R.; Lara, I.; García-Blanco, A.; Cháfer-Pericás, C.; Vento, M. Non-invasive monitoring of stress biomarkers in the newborn period. Semin. Fetal Neonatal Med. 2019, 24, 101002. [Google Scholar] [CrossRef]
- Ho, J.E.; Lyass, A.; Courchesne, P.; Chen, G.; Liu, C.; Yin, X.; Hwang, S.J.; Massaro, J.M.; Larson, M.G.; Levy, D. Protein Biomarkers of Cardiovascular Disease and Mortality in the Community. J. Am. Heart Assoc. 2018, 7, e008108. [Google Scholar] [CrossRef]
- Brankovic, M.; Akkerhuis, K.M.; Mouthaan, H.; Brugts, J.J.; Manintveld, O.C.; Van Ramshorst, J.; Germans, T.; Umans, V.; Boersma, E.; Kardys, I. Cardiometabolic Biomarkers and Their Temporal Patterns Predict Poor Outcome in Chronic Heart Failure (Bio-SHiFT Study). J. Clin. Endocrinol. Metab. 2018, 103, 3954–3964. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, C.; Gil-Redondo, R.; Seco, M.; Barragán, R.; de la Cruz, L.; Cannet, C.; Schäfer, H.; Fang, F.; Diercks, T.; Bizkarguenaga, M.; et al. A molecular signature for the metabolic syndrome by urine metabolomics. Cardiovasc. Diabetol. 2021, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lonardo, A.; Byrne, C.D.; Targher, G. Precision medicine approaches in metabolic disorders and target organ damage: Where are we now, and where are we going? Metab. Target Organ Damage 2021, 1. [Google Scholar] [CrossRef]
- Chen, X. Commercial plasma donation and individual health in impoverished rural China. Health Econ. Rev. 2014, 4, 30. [Google Scholar] [CrossRef][Green Version]
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Alonso, A.; Beaton, A.Z.; Bittencourt, M.S.; Boehme, A.K.; Buxton, A.E.; Carson, A.P.; Commodore-Mensah, Y.; et al. Heart Disease and Stroke Statistics—2022 Update: A Report From the American Heart Association. Circulation 2022, 145, e153–e639. [Google Scholar] [CrossRef]
- Dhingra, R.; Vasan, R.S. Biomarkers in cardiovascular disease: Statistical assessment and section on key novel heart failure biomarkers. Trends Cardiovasc. Med. 2017, 27, 123–133. [Google Scholar] [CrossRef]
- Manson, J.E.; Bassuk, S.S. Biomarkers of cardiovascular disease risk in women. Metabolism 2015, 64, S33–S39. [Google Scholar] [CrossRef]
- Wang, J.; Tan, G.-J.; Han, L.-N.; Bai, Y.-Y.; He, M.; Liu, H.-B. Novel biomarkers for cardiovascular risk prediction. J. Geriatr. Cardiol. 2017, 14, 135–150. [Google Scholar] [CrossRef]
- Hozawa, A.; Folsom, A.R.; Sharrett, A.R.; Chambless, L.E. Absolute and Attributable Risks of Cardiovascular Disease Incidence in Relation to Optimal and Borderline Risk Factors–Comparison of African American with white subjects–Atherosclerosis risk in communities study. Arch. Intern. Med. 2007, 167, 573–579. [Google Scholar] [CrossRef]
- Hu, F.B. Globalization of Diabetes The role of diet, lifestyle, and genes. Diabetes Care 2011, 34, 1249–1257. [Google Scholar] [CrossRef]
- Tuomilehto, J.; Schwarz, P.; Lindström, J. Long-Term Benefits from Lifestyle Interventions for Type 2 Diabetes Prevention Time to expand the efforts. Diabetes Care 2011, 34, S210–S214. [Google Scholar] [CrossRef] [PubMed]
- Buysschaert, M.; Medina, J.L.; Buysschaert, B.; Bergman, M. Definitions (and Current Controversies) of Diabetes and Prediabetes. Curr. Diabetes Rev. 2016, 12, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Dorcely, B.; Katz, K.; Jagannathan, R.; Chiang, S.S.; Oluwadare, B.; Goldberg, I.J.; Bergman, M. Novel biomarkers for prediabetes, diabetes, and associated complications. Diabetes Metab. Syndr. Obes. Targets Ther. 2017, 10, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Bonora, E.; Tuomilehto, J. The Pros and Cons of Diagnosing Diabetes with A1C. Diabetes Care 2011, 34, S184–S190. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Is Cancer a Genetic Disease or a Metabolic Disease? eBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef]
- Gyamfi, J.; Kim, J.; Choi, J. Cancer as a Metabolic Disorder. Int. J. Mol. Sci. 2022, 23, 1155. [Google Scholar] [CrossRef]
- Jurica, M.S.; Mesecar, A.; Heath, P.J.; Shi, W.; Nowak, T.; Stoddard, B.L. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 1998, 6, 195–210. [Google Scholar] [CrossRef]
- Raben, D.M. Phosphofructokinase-2/Fructose Bisphosphatase-2. In Encyclopedia of Biological Chemistry, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 456–458. [Google Scholar] [CrossRef]
- Bartrons, R.; Simon-Molas, H.; Rodríguez-García, A.; Castaño, E.; Navarro-Sabaté, À.; Manzano, A.; Martinez-Outschoorn, U.E. Fructose 2,6-Bisphosphate in Cancer Cell Metabolism. Front. Oncol. 2018, 8, 331. [Google Scholar] [CrossRef]
- Beyoğlu, D.; Idle, J. Metabolic Rewiring and the Characterization of Oncometabolites. Cancers 2021, 13, 2900. [Google Scholar] [CrossRef]
- Kennedy, K.M.; Dewhirst, M.W. Tumor metabolism of lactate: The influence and therapeutic potential for MCT and CD147 regulation. Future Oncol. 2010, 6, 127–148. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, L.; Di, L.-J. Compartmentation of Metabolites in Regulating Epigenomes of Cancer. Mol. Med. 2016, 22, 349–360. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seyfried, T.N.; Mukherjee, P.; Iyikesici, M.S.; Slocum, A.; Kalamian, M.; Spinosa, J.-P.; Chinopoulos, C. Consideration of Ketogenic Metabolic Therapy as a Complementary or Alternative Approach for Managing Breast Cancer. Front. Nutr. 2020, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Valle, S.; Alcalá, S.; Martin-Hijano, L.; Cabezas-Sáinz, P.; Navarro, D.; Muñoz, E.R.; Yuste, L.; Tiwary, K.; Walter, K.; Ruiz-Cañas, L.; et al. Exploiting oxidative phosphorylation to promote the stem and immunoevasive properties of pancreatic cancer stem cells. Nat. Commun. 2020, 11, 5265. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Gowda, G.N.; Raftery, D. Metabolic profiling: Are we en route to better diagnostic tests for cancer? Future Oncol. 2012, 8, 1207–1210. [Google Scholar] [CrossRef]
- Li, J.-J.; Fang, C.-H. C-reactive protein is not only an inflammatory marker but also a direct cause of cardiovascular diseases. Med. Hypotheses 2004, 62, 499–506. [Google Scholar] [CrossRef]
- Castro, A.R.; Silva, S.O.; Soares, S.C. The Use of High Sensitivity C-Reactive Protein in Cardiovascular Disease Detection. J. Pharm. Pharm. Sci. 2018, 21, 496–503. [Google Scholar] [CrossRef]
- Zhu, K.; Knuiman, M.; Divitini, M.; Murray, K.; Lim, E.M.; John, A.S.; Walsh, J.; Hung, J. High-sensitivity cardiac troponin I and risk of cardiovascular disease in an Australianpopulation-based cohort. Heart 2018, 104, 895–903. [Google Scholar] [CrossRef]
- Jeong, S.; Choi, S.; Kim, K.; Kim, S.M.; Lee, G.; Park, S.Y.; Kim, Y.; Son, J.S.; Yun, J.; Park, S.M. Effect of Change in Total Cholesterol Levels on Cardiovascular Disease Among Young Adults. J. Am. Heart Assoc. 2018, 7. [Google Scholar] [CrossRef]
- Colpo, A. LDL Cholesterol: ”Bad” Cholesterol or Bad Science? J. Am. Physicians Surg. 2005, 10, 83. [Google Scholar]
- Kingwell, B.A.; Chapman, M.J.; Kontush, A.; Miller, N.E. HDL-targeted therapies: Progress, failures and future. Nat. Rev. Drug Discov. 2014, 13, 445–464. [Google Scholar] [CrossRef]
- Toth, P. Triglyceride-rich lipoproteins as a causal factor for cardiovascular disease. Vasc. Health Risk Manag. 2016, 12, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Haque, T.; Rahman, S.; Islam, S.; Molla, N.H.; Ali, N. Assessment of the relationship between serum uric acid and glucose levels in healthy, prediabetic and diabetic individuals. Diabetol. Metab. Syndr. 2019, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, Z.; Lei, Z.; Lei, P. CD14: Biology and role in the pathogenesis of disease. Cytokine Growth Factor Rev. 2019, 48, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.-T.; Chang, W.-H.; Kuo, T.-F.; Shen, M.-Y.; Yang, C.-W.; Tien, Y.-J.; Lai, B.-Y.; Chen, Y.-R.; Chang, Y.-C.; Yang, W.-C. Identification of Novel Biomarkers for Pre-diabetic Diagnosis Using a Combinational Approach. Front. Endocrinol. 2021, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Furusyo, N.; Hayashi, J. Glycated albumin and diabetes mellitus. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 5509–5514. [Google Scholar] [CrossRef]
- Hotta, K.; Funahashi, T.; Arita, Y.; Takahashi, M.; Matsuda, M.; Okamoto, Y.; Iwahashi, H.; Kuriyama, H.; Ouchi, N.; Maeda, K.; et al. Plasma Concentrations of a Novel, Adipose-Specific Protein, Adiponectin, in Type 2 Diabetic Patients. Arter. Thromb. Vasc. Biol. 2000, 20, 1595–1599. [Google Scholar] [CrossRef]
- Yang, M.; Soga, T.; Pollard, P.J.; Adam, J. The emerging role of fumarate as an oncometabolite. Front. Oncol. 2012, 2, 85. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Propert, K.J.; Loren, A.W.; Paietta, E.; Sun, Z.; Levine, R.L.; Straley, K.S.; Yen, K.; Patel, J.P.; Agresta, S.; et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood 2013, 121, 4917–4924. [Google Scholar] [CrossRef]
- Cernei, N.; Heger, Z.; Gumulec, J.; Zitka, O.; Masarik, M.; Babula, P.; Eckschlager, T.; Stiborova, M.; Kizek, R.; Adam, V. Sarcosine as a Potential Prostate Cancer Biomarker—A Review. Int. J. Mol. Sci. 2013, 14, 13893–13908. [Google Scholar] [CrossRef]
- Niemi, R.J.; Roine, A.N.; Häkkinen, M.R.; Kumpulainen, P.S.; Keinänen, T.A.; Vepsäläinen, J.J.; Lehtimäki, T.; Oksala, N.K.; Mäenpää, J.U. Urinary Polyamines as Biomarkers for Ovarian Cancer. Int. J. Gynecol. Cancer 2017, 27, 1360–1366. [Google Scholar] [CrossRef]
- Andersen, L.W.; Mackenhauer, J.; Roberts, J.C.; Berg, K.M.; Cocchi, M.N.; Donnino, M.W. Etiology and Therapeutic Approach to Elevated Lactate Levels. Mayo Clin. Proc. 2013, 88, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Huijgen, H.J.; Sanders, G.T.; Koster, R.W.; Vreeken, J.; Bossuyt, P.M. The clinical value of lactate dehydrogenase in serum: A quantitative review. Eur. J. Clin. Chem. Clin. Biochem. 1997, 35, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.J.; Dell, S.D.; Gaston, B.; O’Connor, M.; Marozkina, N.; Manion, M.; Hazucha, M.J.; Leigh, M.W. Nasal Nitric Oxide Measurement in Primary Ciliary Dyskinesia. A Technical Paper on Standardized Testing Protocols. Ann. Am. Thorac. Soc. 2020, 17, e1–e12. [Google Scholar] [CrossRef]
- Niinistö, S.; Takkinen, H.M.; Erlund, I.; Ahonen, S.; Toppari, J.; Ilonen, J.; Veijola, R.; Knip, M.; Vaarala, O.; Virtanen, S.M. Fatty acid status in infancy is associated with the risk of type 1 diabetes-associated autoimmunity. Diabetologia. 2017, 60, 1223–1233. [Google Scholar] [CrossRef]
- Tsoukalas, D.; Fragoulakis, V.; Papakonstantinou, E.; Antonaki, M.; Vozikis, A.; Tsatsakis, A.; Buga, A.M.; Mitroi, M.; Calina, D. Prediction of Autoimmune Diseases by Targeted Metabolomic Assay of Urinary Organic Acids. Metabolites 2020, 10, 502. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef]
- Rodriguez-Coira, J.; Villaseñor, A.; Izquierdo, E.; Huang, M.; Barker-Tejeda, T.C.; Radzikowska, U.; Sokolowska, M.; Barber, D. The Importance of Metabolism for Immune Homeostasis in Allergic Diseases. Front. Immunol. 2021, 12, 2919. [Google Scholar] [CrossRef]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef]
- Tsoukalas, D.; Fragoulakis, V.; Sarandi, E.; Docea, A.O.; Papakonstaninou, E.; Tsilimidos, G.; Anamaterou, C.; Fragkiadaki, P.; Aschner, M.; Tsatsakis, A.; et al. Targeted Metabolomic Analysis of Serum Fatty Acids for the Prediction of Autoimmune Diseases. Front. Mol. Biosci. 2019, 6, 120. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef]
- Liebmann, M.; Hucke, S.; Koch, K.; Eschborn, M.; Ghelman, J.; Chasan, A.I.; Glander, S.; Schädlich, M.; Kuhlencord, M.; Daber, N.M.; et al. Nur77 serves as a molecular brake of the metabolic switch during T cell activation to restrict autoimmunity. Proc. Natl. Acad. Sci. USA 2018, 115, E8017–E8026. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.S.; Ghosh, S.; Newman, S.; Salbaum, J.M. A map of the PGC-1α- and NT-PGC-1α-regulated transcriptional network in brown adipose tissue. Sci. Rep. 2018, 8, 7876. [Google Scholar] [CrossRef] [PubMed]
- Katchman, B.A.; Zhu, M.; Blain Christen, J.; Anderson, K.S. Eccrine Sweat as a Biofluid for Profiling Immune Biomarkers. Proteom. Clin. Appl. 2018, 12, e1800010. [Google Scholar] [CrossRef]
- Yu, Y.; Prassas, I.; Muytjens, C.M.; Diamandis, E.P. Proteomic and peptidomic analysis of human sweat with emphasis on proteolysis. J. Proteom. 2017, 155, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Sempionatto, J.R.; Moon, J.-M.; Wang, J. Touch-Based Fingertip Blood-Free Reliable Glucose Monitoring: Personalized Data Processing for Predicting Blood Glucose Concentrations. ACS Sens. 2021, 6, 1875–1883. [Google Scholar] [CrossRef]
- Ates, H.C.; Brunauer, A.; von Stetten, F.; Urban, G.A.; Güder, F.; Merkoçi, A.; Früh, S.M.; Dincer, C. Integrated Devices for Non-Invasive Diagnostics. Adv. Funct. Mater. 2021, 31, 2010388. [Google Scholar] [CrossRef]
- Chung, M.; Fortunato, G.; Radacsi, N. Wearable flexible sweat sensors for healthcare monitoring: A review. J. R. Soc. Interface 2019, 16, 20190217. [Google Scholar] [CrossRef]
- Bandodkar, A.J.; Gutruf, P.; Choi, J.; Lee, K.; Sekine, Y.; Reeder, J.T.; Jeang, W.J.; Aranyosi, A.J.; Lee, S.P.; Model, J.B.; et al. Battery-free, skin-interfaced microfluidic/electronic systems for simultaneous electrochemical, colorimetric, and volumetric analysis of sweat. Sci. Adv. 2019, 5, eaav3294. [Google Scholar] [CrossRef]
- Choi, D.-H.; Kitchen, G.; Kim, J.S.; Li, Y.; Kim, K.; Jeong, I.C.; Nguyen, J.; Stewart, K.J.; Zeger, S.L.; Searson, P.C. Two Distinct Types of Sweat Profile in Healthy Subjects While Exercising at Constant Power Output Measured by a Wearable Sweat Sensor. Sci. Rep. 2019, 9, 17877. [Google Scholar] [CrossRef]
- Nyein, H.Y.Y.; Tai, L.-C.; Ngo, Q.P.; Chao, M.; Zhang, G.B.; Gao, W.; Bariya, M.; Bullock, J.; Kim, H.; Fahad, H.M.; et al. A Wearable Microfluidic Sensing Patch for Dynamic Sweat Secretion Analysis. ACS Sens. 2018, 3, 944–952. [Google Scholar] [CrossRef]
- Yang, Y.; Song, Y.; Bo, X.; Min, J.; Pak, O.S.; Zhu, L.; Wang, M.; Tu, J.; Kogan, A.; Zhang, H.; et al. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 2020, 38, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yang, Z.; An, M. Lab on the eye: A review of tear-based wearable devices for medical use and health management. Biosci. Trends 2019, 13, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Hagan, S.; Tomlinson, A.; Madden, L.; Clark, A.M.; Oliver, K. Analysis of tear fluid proteins: Use of multiplex assays in profiling biomarkers of dry eye disease. EPMA J. 2014, 5, A159. [Google Scholar] [CrossRef][Green Version]
- Kalló, G.; Emri, M.; Varga, Z.; Ujhelyi, B.; Tőzsér, J.; Csutak, A.; Csősz, E. Changes in the Chemical Barrier Composition of Tears in Alzheimer’s Disease Reveal Potential Tear Diagnostic Biomarkers. PLoS ONE 2016, 11, e0158000. [Google Scholar] [CrossRef]
- Wu, J.; Sigler, A.; Pfaff, A.; Cen, N.; Ercal, N.; Shi, H. Development of a HPLC-MS/MS method for assessment of thiol redox status in human tear fluids. Anal. Biochem. 2021, 629, 114295. [Google Scholar] [CrossRef]
- Lam, S.M.; Tong, L.; Reux, B.; Duan, X.; Petznick, A.; Yong, S.S.; Khee, C.B.S.; Lear, M.J.; Wenk, M.R.; Shui, G. Lipidomic analysis of human tear fluid reveals structure-specific lipid alterations in dry eye syndrome. J. Lipid Res. 2014, 55, 299–306. [Google Scholar] [CrossRef]
- Kenny, A.; Jiménez-Mateos, E.M.; Zea-Sevilla, M.A.; Rábano, A.; Gili-Manzanaro, P.; Prehn, J.H.M.; Henshall, D.C.; Ávila, J.; Engel, T.; Hernández, F. Proteins and microRNAs are differentially expressed in tear fluid from patients with Alzheimer’s disease. Sci. Rep. 2019, 9, 15437. [Google Scholar] [CrossRef]
- Brunmair, J.; Bileck, A.; Schmidl, D.; Hagn, G.; Meier-Menches, S.M.; Hommer, N.; Schlatter, A.; Gerner, C.; Garhöfer, G. Metabolic phenotyping of tear fluid as a prognostic tool for personalised medicine exemplified by T2DM patients. EPMA J. 2022, 13, 107–123. [Google Scholar] [CrossRef]
- Brunauer, A.; Ates, H.C.; Dincer, C.; Früh, S.M. Integrated paper-based sensing devices for diagnostic applications. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 89, pp. 397–450. [Google Scholar]
- Das, S.; Pal, M. Review—Non-Invasive Monitoring of Human Health by Exhaled Breath Analysis: A Comprehensive Review. J. Electrochem. Soc. 2020, 167, 037562. [Google Scholar] [CrossRef]
- Chen, T.; Liu, T.; Li, T.; Zhao, H.; Chen, Q. Exhaled breath analysis in disease detection. Clin. Chim. Acta 2021, 515, 61–72. [Google Scholar] [CrossRef]
- Kalapos, M. Possible physiological roles of acetone metabolism in humans. Med. Hypotheses 1999, 53, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Righettoni, M.; Tricoli, A.; Pratsinis, S.E. Si:WO3Sensors for Highly Selective Detection of Acetone for Easy Diagnosis of Diabetes by Breath Analysis. Anal. Chem. 2010, 82, 3581–3587. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.-J.; Suzuki, T.; Tsujii, M.; Ye, M.; Minami, I.; Toda, K.; Otsuka, H.; Toma, K.; Arakawa, T.; Araki, K.; et al. Biochemical Gas Sensors (Biosniffers) Using Forward and Reverse Reactions of Secondary Alcohol Dehydrogenase for Breath Isopropanol and Acetone as Potential Volatile Biomarkers of Diabetes Mellitus. Anal. Chem. 2017, 89, 12261–12268. [Google Scholar] [CrossRef]
- Gould, O.; Ratcliffe, N.; Król, E.; Costello, B.D.L. Breath analysis for detection of viral infection, the current position of the field. J. Breath Res. 2020, 14, 041001. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, D.R.; Li, R.T.; Voos, J.E.; Rowbottom, J.R.; Alfes, C.M.; Zorman, C.A.; Drummond, C.K. Wearable sensors for monitoring the physiological and biochemical profile of the athlete. Digit. Med. 2019, 2, 72. [Google Scholar] [CrossRef] [PubMed]
- Nunes, L.; Mussavira, S.; Bindhu, O.S. Clinical and diagnostic utility of saliva as a non-invasive diagnostic fluid: A systematic review. Biochem. Med. 2015, 25, 177–192. [Google Scholar] [CrossRef]
- Desai, P.; Donovan, L.; Janowitz, E.; Kim, J.Y. The Clinical Utility of Salivary Biomarkers in the Identification of Type 2 Diabetes Risk and Metabolic Syndrome. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 3587–3599. [Google Scholar] [CrossRef]
- Kim, J.; Valdés-Ramírez, G.; Bandodkar, A.J.; Jia, W.; Martinez, A.G.; Ramírez, J.; Mercier, P.; Wang, J. Non-invasive mouthguard biosensor for continuous salivary monitoring of metabolites. Analyst 2014, 139, 1632–1636. [Google Scholar] [CrossRef]
- García-Carmona, L.; Martín, A.; Sempionatto, J.R.; Moreto, J.R.; González, M.C.; Wang, J.; Escarpa, A. Pacifier Biosensor: Toward Noninvasive Saliva Biomarker Monitoring. Anal. Chem. 2019, 91, 13883–13891. [Google Scholar] [CrossRef]
- Kim, J.; Imani, S.; de Araujo, W.R.; Warchall, J.; Valdés-Ramírez, G.; Paixao, T.R.L.C.; Mercier, P.P.; Wang, J. Wearable salivary uric acid mouthguard biosensor with integrated wireless electronics. Biosens. Bioelectron. 2015, 74, 1061–1068. [Google Scholar] [CrossRef]
- Low, S.S.; Pan, Y.; Ji, D.; Li, Y.; Lu, Y.; He, Y.; Chen, Q.; Liu, Q. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens. Actuators B Chem. 2020, 308, 127718. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K.; Prenzler, P.; Kendall, M. Recent and potential developments in the analysis of urine: A review. Anal. Chim. Acta 2011, 684, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Harpole, M.; Davis, J.; Espina, V. Current state of the art for enhancing urine biomarker discovery. Expert Rev. Proteom. 2016, 13, 609–626. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, G.A.; Cass, W.A.; Hudson, J.; Henson, M.; Zhang, Z.; Ovadia, A.; Hoffer, B.J.; Gash, D.M. In Vivo Electrochemical Studies of Dopamine Overflow and Clearance in the Striatum of Normal and MPTP-Treated Rhesus Monkeys. J. Neurochem. 1996, 66, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Garris, P.A.; Christensen, J.R.C.; Rebec, G.V.; Wightman, R.M. Real-Time Measurement of Electrically Evoked Extracellular Dopamine in the Striatum of Freely Moving Rats. J. Neurochem. 1997, 68, 152–161. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Nolan, J.K.; Park, H.; Lam, S.; Fattah, M.; Page, J.C.; Joe, H.-E.; Jun, M.; Lee, H.; Kim, S.J.; et al. Corrigendum to Facile fabrication of flexible glutamate biosensor using direct writing of platinum nanoparticle-based nanocomposite ink. Biosens. Bioelectron. 2019, 141, 111429. [Google Scholar] [CrossRef]
- Choo, S.-S.; Kang, E.-S.; Song, I.; Lee, D.; Choi, J.-W.; Kim, T.-H. Electrochemical Detection of Dopamine Using 3D Porous Graphene Oxide/Gold Nanoparticle Composites. Sensors 2017, 17, 861. [Google Scholar] [CrossRef]
- Sáenz, H.S.C.; Hernández-Saravia, L.P.; Selva, J.S.G.; Sukeri, A.; Espinoza-Montero, P.J.; Bertotti, M. Electrochemical dopamine sensor using a nanoporous gold microelectrode: A proof-of-concept study for the detection of dopamine release by scanning electrochemical microscopy. Mikrochim. Acta 2018, 185, 367. [Google Scholar] [CrossRef]
- Sheng, Z.-H.; Zheng, X.-Q.; Xu, J.-Y.; Bao, W.-J.; Wang, F.-B.; Xia, X.-H. Electrochemical sensor based on nitrogen doped graphene: Simultaneous determination of ascorbic acid, dopamine and uric acid. Biosens. Bioelectron. 2012, 34, 125–131. [Google Scholar] [CrossRef]
- Burmeister, J.J.; Price, D.A.; Pomerleau, F.; Huettl, P.; Quintero, J.E.; Gerhardt, G.A. Challenges of simultaneous measurements of brain extracellular GABA and glutamate in vivo using enzyme-coated microelectrode arrays. J. Neurosci. Methods 2020, 329, 108435. [Google Scholar] [CrossRef]
- Hossain, I.; Tan, C.; Doughty, P.T.; Dutta, G.; Murray, T.A.; Siddiqui, S.; Iasemidis, L.; Arumugam, P.U. A Novel Microbiosensor Microarray for Continuous ex Vivo Monitoring of Gamma-Aminobutyric Acid in Real-Time. Front. Neurosci. 2018, 12, 500. [Google Scholar] [CrossRef] [PubMed]
- Doughty, P.T.; Hossain, I.; Gong, C.; Ponder, K.A.; Pati, S.; Arumugam, P.U.; Murray, T.A. Novel microwire-based biosensor probe for simultaneous real-time measurement of glutamate and GABA dynamics in vitro and in vivo. Sci. Rep. 2020, 10, 12777. [Google Scholar] [CrossRef] [PubMed]
- Boonkaew, S.; Jang, I.; Noviana, E.; Siangproh, W.; Chailapakul, O.; Henry, C.S. Electrochemical paper-based analytical device for multiplexed, point-of-care detection of cardiovascular disease biomarkers. Sens. Actuators B Chem. 2021, 330, 129336. [Google Scholar] [CrossRef]
- Koukouviti, E.; Kokkinos, C. 3D printed enzymatic microchip for multiplexed electrochemical biosensing. Anal. Chim. Acta 2021, 1186, 339114. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, B.; Reeder, J.T.; Seo, S.H.; Lee, S.-U.; Hourlier-Fargette, A.; Shin, J.; Sekine, Y.; Jeong, H.; Oh, Y.S.; et al. Soft, skin-interfaced microfluidic systems with integrated immunoassays, fluorometric sensors, and impedance measurement capabilities. Proc. Natl. Acad. Sci. USA 2020, 117, 27906–27915. [Google Scholar] [CrossRef]
- Tang, C.K.; Vaze, A.; Shen, M.; Rusling, J.F. High-Throughput Electrochemical Microfluidic Immunoarray for Multiplexed Detection of Cancer Biomarker Proteins. ACS Sens. 2016, 1, 1036–1043. [Google Scholar] [CrossRef]
- Xu, T.; Song, Y.; Gao, W.; Wu, T.; Xu, L.-P.; Zhang, X.; Wang, S. Superwettable Electrochemical Biosensor toward Detection of Cancer Biomarkers. ACS Sens. 2018, 3, 72–78. [Google Scholar] [CrossRef]
- Gao, W.; Emaminejad, S.; Nyein, H.Y.Y.; Challa, S.; Chen, K.; Peck, A.; Fahad, H.M.; Ota, H.; Shiraki, H.; Kiriya, D.; et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 2016, 529, 509–514. [Google Scholar] [CrossRef]
- Chinnadayyala, S.R.; Cho, S. Electrochemical Immunosensor for the Early Detection of Rheumatoid Arthritis Biomarker: Anti-Cyclic Citrullinated Peptide Antibody in Human Serum Based on Avidin-Biotin System. Sensors 2021, 21, 124. [Google Scholar] [CrossRef]
- Ren, X.; Ellis, B.W.; Ronan, G.; Blood, S.; DeShetler, C.; Senapati, S.; March, K.L.; Handberg, E.; Anderson, D.; Pepine, C.; et al. A multiplexed ion-exchange membrane-based miRNA (MIX·miR) detection platform for rapid diagnosis of myocardial infarction. Lab Chip 2021, 21, 3876–3887. [Google Scholar] [CrossRef]
- Vargas, E.; Teymourian, H.; Tehrani, F.; Eksin, E.; Sánchez-Tirado, E.; Warren, P.; Erdem, A.; Dassau, E.; Wang, J. Enzymatic/Immunoassay Dual-Biomarker Sensing Chip: Towards Decentralized Insulin/Glucose Detection. Angew. Chem. Int. Ed. 2019, 58, 6376–6379. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Nayak, H.C.; Rajput, S.; Roy, S.; Nagabooshanam, S.; Wadhwa, S.; Kumar, R. An Enzymatic Multiplexed Impedimetric Sensor Based on α-MnO2/GQD Nano-Composite for the Detection of Diabetes and Diabetic Foot Ulcer Using Micro-Fluidic Platform. Chemosensors 2021, 9, 339. [Google Scholar] [CrossRef]
- Zhai, S.; Fang, C.; Yan, J.; Zhao, Q.; Tu, Y. A label-free genetic biosensor for diabetes based on AuNPs decorated ITO with electrochemiluminescent signaling. Anal. Chim. Acta 2017, 982, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Shu, J.; Ju, P.; Wu, S.; Chen, Y.; Ding, S.; Yang, W.; Luo, R. A stochastic DNA walker electrochemiluminescence biosensor based on quenching effect of Pt@CuS on luminol@Au/Ni-Co nanocages for ultrasensitive detection of I27L gene. Chem. Eng. J. 2022, 434, 134681. [Google Scholar] [CrossRef]
- Ngamdee, T.; Chalermwatanachai, T.; Siriwan, C.; Warachit, O.; Rijiravanich, P.; Surareungchai, W. Target amplification-free detection of urinary microRNA for diabetic nephropathy diagnosis with electrocatalytic reaction. Anal. Bioanal. Chem. 2022, 414, 5695–5707. [Google Scholar] [CrossRef]
- Smith, D.A.; Simpson, K.; Cicero, M.L.; Newbury, L.J.; Nicholas, P.; Fraser, D.J.; Caiger, N.; Redman, J.E.; Bowen, T. Detection of urinary microRNA biomarkers using diazo sulfonamide-modified screen printed carbon electrodes. RSC Adv. 2021, 11, 18832–18839. [Google Scholar] [CrossRef]
- Cao, Z.; Duan, F.; Huang, X.; Liu, Y.; Zhou, N.; Xia, L.; Zhang, Z.; Du, M. A multiple aptasensor for ultrasensitive detection of miRNAs by using covalent-organic framework nanowire as platform and shell-encoded gold nanoparticles as signal labels. Anal. Chim. Acta 2019, 1082, 176–185. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, X.; Yang, J.; He, Y.; Li, Y. Application of Multiplex Microfluidic Electrochemical Sensors in Monitoring Hematological Tumor Biomarkers. Anal. Chem. 2020, 92, 11981–11986. [Google Scholar] [CrossRef]
- Pimalai, D.; Putnin, T.; Waiwinya, W.; Chotsuwan, C.; Aroonyadet, N.; Japrung, D. Development of electrochemical biosensors for simultaneous multiplex detection of microRNA for breast cancer screening. Mikrochim. Acta 2021, 188, 329. [Google Scholar] [CrossRef]
- Pothipor, C.; Jakmunee, J.; Bamrungsap, S.; Ounnunkad, K. An electrochemical biosensor for simultaneous detection of breast cancer clinically related microRNAs based on a gold nanoparticles/graphene quantum dots/graphene oxide film. Analyst 2021, 146, 4000–4009. [Google Scholar] [CrossRef]
- Guerrero, S.; Sánchez-Tirado, E.; Agüí, L.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J. Simultaneous determination of CXCL7 chemokine and MMP3 metalloproteinase as biomarkers for rheumatoid arthritis. Talanta 2021, 234, 122705. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, B.; Blázquez-García, M.; Valverde, A.; Serafín, V.; Montero-Calle, A.; Solís-Fernández, G.; Barderas, R.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Binary MoS2 nanostructures as nanocarriers for amplification in multiplexed electrochemical immunosensing: Simultaneous determination of B cell activation factor and proliferation-induced signal immunity-related cytokines. Mikrochim. Acta 2022, 189, 143. [Google Scholar] [CrossRef]
- Tabassum, S.; Kumar, R. Advances in Fiber-Optic Technology for Point-of-Care Diagnosis and In Vivo Biosensing. Adv. Mater. Technol. 2020, 5, 1900792. [Google Scholar] [CrossRef]
- Latifi, H.; Zibaii, M.I.; Hosseini, S.M.; Jorge, P. Nonadiabatic tapered optical fiber for biosensor applications. Photonic Sens. 2012, 2, 340–356. [Google Scholar] [CrossRef]
- Ni, Y.; Rosier, B.J.H.M.; van Aalen, E.A.; Hanckmann, E.T.L.; Biewenga, L.; Pistikou, A.-M.M.; Timmermans, B.; Vu, C.; Roos, S.; Arts, R.; et al. A plug-and-play platform of ratiometric bioluminescent sensors for homogeneous immunoassays. Nat. Commun. 2021, 12, 4586. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-H.; Pan, M.-J.; Jargalsaikhan, Z.; Ishdorj, T.-O.; Tseng, F.-G. Development of Surface-Enhanced Raman Scattering (SERS)-Based Surface-Corrugated Nanopillars for Biomolecular Detection of Colorectal Cancer. Biosensors 2020, 10, 163. [Google Scholar] [CrossRef]
- Balboni, I.; Limb, C.; Tenenbaum, J.; Utz, P. Sa.21. Evaluation of Microarray Surfaces and Arraying Parameters for Autoantibody Profiling. Clin. Immunol. 2008, 127, S87. [Google Scholar] [CrossRef]
- Spindel, S.; Sapsford, K.E. Evaluation of Optical Detection Platforms for Multiplexed Detection of Proteins and the Need for Point-of-Care Biosensors for Clinical Use. Sensors 2014, 14, 22313–22341. [Google Scholar] [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef]
- Righetti, P.G.; Campostrini, N.; Pascali, J.; Hamdan, M.; Astner, H. Quantitative Proteomics: A Review of Different Methodologies. Eur. J. Mass Spectrom. 2004, 10, 335–348. [Google Scholar] [CrossRef]
- Oleinikov, V.A. Fluorescent semiconductor nanocrystals (quantum dots) in protein biochips. Russ. J. Bioorg. Chem. 2011, 37, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Kubina, R.; Dziedzic, A. Molecular and Serological Tests for COVID-19 a Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Jahns, S.; Bräu, M.; Meyer, B.-O.; Karrock, T.; Gutekunst, S.B.; Blohm, L.; Selhuber-Unkel, C.; Buhmann, R.; Nazirizadeh, Y.; Gerken, M. Handheld imaging photonic crystal biosensor for multiplexed, label-free protein detection. Biomed. Opt. Express 2015, 6, 3724–3736. [Google Scholar] [CrossRef] [PubMed]
- Bläsi, J.; Gerken, M. Multiplex optical biosensors based on multi-pinhole interferometry. Biomed. Opt. Express 2021, 12, 4265–4275. [Google Scholar] [CrossRef] [PubMed]
- Qavi, A.J.; Kindt, J.T.; Gleeson, M.A.; Bailey, R.C. Anti-DNA:RNA Antibodies and Silicon Photonic Microring Resonators: Increased Sensitivity for Multiplexed microRNA Detection. Anal. Chem. 2011, 83, 5949–5956. [Google Scholar] [CrossRef]
- Chen, P.; Chung, M.T.; McHugh, W.; Nidetz, R.; Li, Y.; Fu, J.; Cornell, T.T.; Shanley, T.P.; Kurabayashi, K. Multiplex Serum Cytokine Immunoassay Using Nanoplasmonic Biosensor Microarrays. ACS Nano 2015, 9, 4173–4181. [Google Scholar] [CrossRef]
- Fang, S.; Li, B.; Song, D.; Zhang, J.; Sun, W.; Yuan, L. A Smart Graded-index Multimode Fiber Based Sensor Unit for Multi-parameter Sensing Applications. Opt. Photon. J. 2013, 3, 265–267. [Google Scholar] [CrossRef]
- Qu, J.-H.; Peeters, B.; Delport, F.; Vanhoorelbeke, K.; Lammertyn, J.; Spasic, D. Gold nanoparticle enhanced multiplexed biosensing on a fiber optic surface plasmon resonance probe. Biosens. Bioelectron. 2021, 192, 113549. [Google Scholar] [CrossRef]
- Rosman, C.; Prasad, J.; Neiser, A.; Henkel, A.; Edgar, J.; Sönnichsen, C. Multiplexed Plasmon Sensor for Rapid Label-Free Analyte Detection. Nano Lett. 2013, 13, 3243–3247. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Song, X.; Wang, H.; Wang, J.; Wang, Y.; Huang, J.; Yu, J. Robust and Universal SERS Sensing Platform for Multiplexed Detection of Alzheimer’s Disease Core Biomarkers Using PAapt-AuNPs Conjugates. ACS Sens. 2019, 4, 2140–2149. [Google Scholar] [CrossRef]
- Mu, Z.; Zhao, X.; Huang, Y.; Lu, M.; Gu, Z. Photonic Crystal Hydrogel Enhanced Plasmonic Staining for Multiplexed Protein Analysis. Small 2015, 11, 6036–6043. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Liu, Z.; Hu, J.; Gunawardena, D.; Tam, H.-Y. Optofluidics in Microstructured Optical Fibers. Micromachines 2018, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Loke, G.; Fink, Y.; Anikeeva, P. Flexible fiber-based optoelectronics for neural interfaces. Chem. Soc. Rev. 2019, 48, 1826–1852. [Google Scholar] [CrossRef]
- Bayindir, M.; Abouraddy, A.F.; Arnold, J.; Joannopoulos, J.D.; Fink, Y. Thermal-Sensing Fiber Devices by Multimaterial Codrawing. Adv. Mater. 2006, 18, 845–849. [Google Scholar] [CrossRef]
- Stolyarov, A.M.; Gumennik, A.; McDaniel, W.; Shapira, O.; Schell, B.; Sorin, F.; Kuriki, K.; Benoit, G.; Rose, A.; Joannopoulos, J.D.; et al. Enhanced chemiluminescent detection scheme for trace vapor sensing in pneumatically-tuned hollow core photonic bandgap fibers. Opt. Express 2012, 20, 12407–12415. [Google Scholar] [CrossRef]
- Gumennik, A.; Stolyarov, A.M.; Schell, B.R.; Hou, C.; Lestoquoy, G.; Sorin, F.; McDaniel, W.; Rose, A.; Joannopoulos, J.D.; Fink, Y. All-in-Fiber Chemical Sensing. Adv. Mater. 2012, 24, 6005–6009. [Google Scholar] [CrossRef]
- Minev, I.R.; Musienko, P.; Hirsch, A.; Barraud, Q.; Wenger, N.; Moraud, E.M.; Gandar, J.; Capogrosso, M.; Milekovic, T.; Asboth, L.; et al. Electronic dura mater for long-term multimodal neural interfaces. Science 2015, 347, 159–163. [Google Scholar] [CrossRef]
- Rein, M.; Levy, E.; Gumennik, A.; Abouraddy, A.F.; Joannopoulos, J.; Fink, Y. Self-assembled fibre optoelectronics with discrete translational symmetry. Nat. Commun. 2016, 7, 12807. [Google Scholar] [CrossRef]
- Abouraddy, A.F.; Bayindir, M.; Benoit, G.; Hart, S.D.; Kuriki, K.; Orf, N.; Shapira, O.; Sorin, F.; Temelkuran, B.; Fink, Y. Towards multimaterial multifunctional fibres that see, hear, sense and communicate. Nat. Mater. 2007, 6, 336–347. [Google Scholar] [CrossRef]
- Rein, M.; Favrod, V.D.; Hou, C.; Khudiyev, T.; Stolyarov, A.; Cox, J.; Chung, C.-C.; Chhav, C.; Ellis, M.; Joannopoulos, J.; et al. Diode fibres for fabric-based optical communications. Nature 2018, 560, 214–218. [Google Scholar] [CrossRef]
- Kundu, S.; Tabassum, S.; Kumar, R. Plasmonic Point-of-Care Device for Sepsis Biomarker Detection. IEEE Sens. J. 2021, 21, 18837–18846. [Google Scholar] [CrossRef]
- Masson, J.-F.; Battaglia, T.M.; Khairallah, P.; Beaudoin, S.; Booksh, K.S. Quantitative Measurement of Cardiac Markers in Undiluted Serum. Anal. Chem. 2007, 79, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Wang, C.-W. Highly specific detection of interleukin-6 (IL-6) protein using combination tapered fiber-optic biosensor dip-probe. Biosens. Bioelectron. 2009, 24, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Kang, K.A. Preliminary Study of Fiber Optic Multi-Cardiac-Marker Biosensing System for Rapid Coronary Heart Disease Diagnosis and Prognosis. Adv. Exp. Med. Biol. 2006, 578, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Noushin, T.; Tabassum, S. WRRIST: A wearable, rapid, and real-time infection screening tool for dual-mode detection of inflammatory biomarkers in sweat. In Proceedings of the Microfluidics, BioMEMS, and Medical Microsystems XX, SPIE BiOS, San Francisco, CA, USA, 22–27 January 2022; p. 1195502. [Google Scholar]
- Aliberti, A.; Ricciardi, A.; Giaquinto, M.; Micco, A.; Bobeico, E.; La Ferrara, V.; Ruvo, M.; Cutolo, A.; Cusano, A. Microgel assisted Lab-on-Fiber Optrode. Sci. Rep. 2017, 7, 14459. [Google Scholar] [CrossRef]
- Koman, V.B.; Santschi, C.; Martin, O.J.F. Multiscattering-enhanced optical biosensor: Multiplexed, non-invasive and continuous measurements of cellular processes. Biomed. Opt. Express 2015, 6, 2353–2365. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Kim, S. Multiplexed detection of biomolecules using a wax printed paper-disc centrifugal optical device. Sens. Actuators B Chem. 2020, 303, 127195. [Google Scholar] [CrossRef]
- Sanders, M.; Lin, Y.; Wei, J.; Bono, T.; Lindquist, R.G. An enhanced LSPR fiber-optic nanoprobe for ultrasensitive detection of protein biomarkers. Biosens. Bioelectron. 2014, 61, 95–101. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Blackie, E.; Meyer, M.; Etchegoin, P.G. Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. J. Phys. Chem. C 2007, 111, 13794–13803. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Wang, F.; Jia, Z.; Zhou, J.; Jiang, T.; Petti, L.; Chen, Y.; Xiong, Q.; Wang, X. Classification analyses for prostate cancer, benign prostate hyperplasia and healthy subjects by SERS-based immunoassay of multiple tumour markers. Talanta 2018, 188, 238–244. [Google Scholar] [CrossRef]
- Chen, R.; Liu, B.; Ni, H.; Chang, N.; Luan, C.; Ge, Q.; Dong, J.; Zhao, X. Vertical flow assays based on core–shell SERS nanotags for multiplex prostate cancer biomarker detection. Analyst 2019, 144, 4051–4059. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Lu, L.; Rong, Z.; Wang, C.; Peng, Y.; Wang, F.; Wang, J.; Sun, M.; Dong, J.; Wang, D.; et al. Portable and multiplexed lateral flow immunoassay reader based on SERS for highly sensitive point-of-care testing. Biosens. Bioelectron. 2020, 168, 112524. [Google Scholar] [CrossRef] [PubMed]
- Aidoo-Brown, J.; Moschou, D.; Estrela, P. Multiplexed Prostate Cancer Companion Diagnostic Devices. Sensors 2021, 21, 5023. [Google Scholar] [CrossRef]
- Washburn, A.L.; Shia, W.W.; Lenkeit, K.A.; Lee, S.-H.; Bailey, R.C. Multiplexed cancer biomarker detection using chip-integrated silicon photonic sensor arrays. Analyst 2016, 141, 5358–5365. [Google Scholar] [CrossRef]
- Huang, R.; Liao, Y.; Zhou, X.; Fu, Y.; Xing, D. Multiplexed detection of microRNA biomarkers from tumor cells and tissues with a homogeneous nano-photon switch. Sens. Actuators Chem. 2017, 247, 505–513. [Google Scholar] [CrossRef]
- Xue, T.; Liang, W.; Li, Y.; Sun, Y.; Xiang, Y.; Zhang, Y.; Dai, Z.; Duo, Y.; Wu, L.; Qi, K.; et al. Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor. Nat. Commun. 2019, 10, 28. [Google Scholar] [CrossRef]
- Graybill, R.M.; Para, C.S.; Bailey, R.C. PCR-Free, Multiplexed Expression Profiling of microRNAs Using Silicon Photonic Microring Resonators. Anal. Chem. 2016, 88, 10347–10351. [Google Scholar] [CrossRef]
- Qavi, A.J.; Bailey, R.C. Multiplexed Detection and Label-Free Quantitation of MicroRNAs Using Arrays of Silicon Photonic Microring Resonators. Angew. Chem. Int. Ed. 2010, 49, 4608–4611. [Google Scholar] [CrossRef]
- Eltzov, E.; Cosnier, S.; Marks, R. Biosensors based on combined optical and electrochemical transduction for molecular diagnostics. Expert Rev. Mol. Diagn. 2011, 11, 533–546. [Google Scholar] [CrossRef]
- Juan-Colás, J.; Johnson, S.; Krauss, T.F. Dual-Mode Electro-Optical Techniques for Biosensing Applications: A Review. Sensors 2017, 17, 2047. [Google Scholar] [CrossRef]
- Dulay, S.; Rivas, L.; Pla, L.; Berdún, S.; Eixarch, E.; Gratacós, E.; Illa, M.; Mir, M.; Samitier, J. Fetal ischemia monitoring with in vivo implanted electrochemical multiparametric microsensors. J. Biol. Eng. 2021, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wu, F.; Yu, P.; Mao, L. In Vivo Electrochemical Sensors for Neurochemicals: Recent Update. ACS Sens. 2019, 4, 3102–3118. [Google Scholar] [CrossRef] [PubMed]
- Dulay, S.; Rivas, L.; Miserere, S.; Pla, L.; Berdún, S.; Parra, J.; Eixarch, E.; Gratacós, E.; Illa, M.; Mir, M.; et al. in vivo Monitoring with micro-implantable hypoxia sensor based on tissue acidosis. Talanta 2021, 226, 122045. [Google Scholar] [CrossRef] [PubMed]
- Vadgama, P. Monitoring with In Vivo Electrochemical Sensors: Navigating the Complexities of Blood and Tissue Reactivity. Sensors 2020, 20, 3149. [Google Scholar] [CrossRef] [PubMed]
- Poeggel, S.; Duraibabu, D.; Tosi, D.; Leen, G.; Lewis, E.; McGrath, D.; Fusco, F.; Sannino, S.; Lupoli, L.; Ippolito, J.; et al. Differential in vivo urodynamic measurement in a single thin catheter based on two optical fiber pressure sensors. J. Biomed. Opt. 2015, 20, 037005. [Google Scholar] [CrossRef]
- Desmet, C.; Vindas, K.; Meza, R.A.; Garrigue, P.; Voci, S.; Sojic, N.; Maziz, A.; Courson, R.; Malaquin, L.; Leichle, T.; et al. Multiplexed Remote SPR Detection of Biological Interactions through Optical Fiber Bundles. Sensors 2020, 20, 511. [Google Scholar] [CrossRef]
- Swetha, P.D.P.; Sonia, J.; Sapna, K.; Prasad, K.S. Towards CRISPR powered electrochemical sensing for smart diagnostics. Curr. Opin. Electrochem. 2021, 30, 100829. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Newsham, E.; Richards-Kortum, R. CRISPR-Based Electrochemical Sensor Permits Sensitive and Specific Viral Detection in Low-Resource Settings. ACS Cent. Sci. 2021, 7, 926–928. [Google Scholar] [CrossRef]
- Chang, C.-H.; Lin, C.-H.; Lane, H.-Y. Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2761. [Google Scholar] [CrossRef]
- Rehman, O.; Zhuang, H.; Ali, A.M.; Ibrahim, A.; Li, Z. Validation of miRNAs as Breast Cancer Biomarkers with a Machine Learning Approach. Cancers 2019, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Tabl, A.A.; Alkhateeb, A.; Elmaraghy, W.; Rueda, L.; Ngom, A. A Machine Learning Approach for Identifying Gene Biomarkers Guiding the Treatment of Breast Cancer. Front. Genet. 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Jagga, Z.; Gupta, D. Machine learning for biomarker identification in cancer research–developments toward its clinical application. Pers. Med. 2015, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Tabassum, S.; Wang, Q.; Wang, Y.; Kumar, R.; Dong, L. Integrated dual-modality microfluidic sensor for biomarker detection using lithographic plasmonic crystal. Lab Chip 2018, 18, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, F.; Brigo, L.; Favaro, M.; Luni, C.; Zoso, A.; Cattelan, M.; Agnoli, S.; Brusatin, G.; Granozzi, G.; Giomo, M.; et al. Optoelectrochemical Biorecognition by Optically Transparent Highly Conductive Graphene-Modified Fluorine-Doped Tin Oxide Substrates. ACS Appl. Mater. Interfaces 2014, 6, 22769–22777. [Google Scholar] [CrossRef]


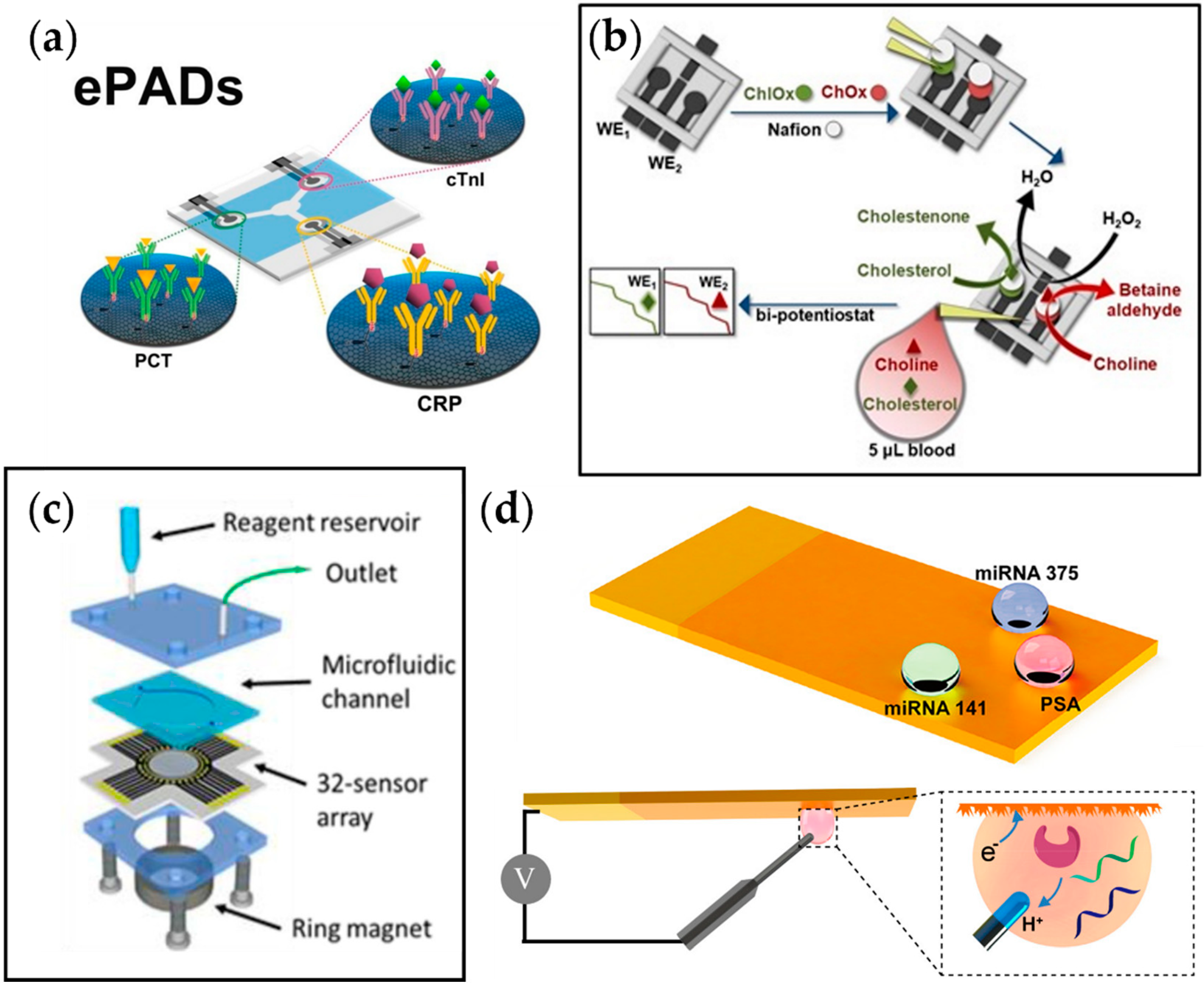
| Metabolic Syndrome | Biomarker | E-Chem Method | LOD | Linear Range | Refs. |
|---|---|---|---|---|---|
| Cardiovascular Diseases (CVDs) | C-reactive protein (CRP) | SWV | 0.38 ng mL−1 | 1–10,000 ng mL−1 | [106] |
| Troponin (cTnI) | 0.16 pg mL−1 | 0.001–250 ng mL−1 | |||
| Procalcitonin (PCT) | 0.27 pg mL−1 | 0.0005–250 ng mL−1 | |||
| Cholesterol | Amperometry | 0.36 μmol L−1 | 30–240 μmol L−1 | [107] | |
| Choline | 0.08 μmol L−1 | 0.5–4 μmol L−1 | |||
| miR-1 | 0.31 pM | ||||
| miR-208b | EIS | 0.37 pM | 0.1 pM–10 nM | [113] | |
| miR-499 | 0.77 pM | ||||
| Diabetes | Glucose | Amperometry | 209 μmol | [114] | |
| Insulin | 340 μmol | ||||
| Glucose | Amperometry | 0–200 μM | [111] | ||
| Lactate | 0–30 μM | ||||
| Glucose | EIS | 58 mg dL−1 | 50–800 mg dL−1 | [115] | |
| L-tyrosine | 0.3 μmol L−1 | 1–500 μmol L−1 | |||
| I27L | ECL | 8.1 × 10−12 M | 1.0 × 10−11–1.0 × 10−7 M | [116] | |
| I27L | ECL | 23 fM | 0.0001–100 nM | [117] | |
| miRNA-124 | DPV | 0.65 fM | 1 fM–100 nM | [118] | |
| miRNA-21 | coulometry | 17 fM | 10−8–10−14 M | [119] | |
| Cancer | miRNA-155 | DPV | 6.7 fM | 0.01–1000 pM | [120] |
| miRNA-122 | 1.5 fM | 0.01–1000 pM | |||
| Prostate Cancer | PSA | DPV | 1–100,000 pg mL−1 | [110] | |
| PSMA Interleukin-6 (IL-6) | 1–10,000 pg mL−1 | ||||
| 1–1000 pg mL−1 | |||||
| Platelet factor-4 (PF-4) | 1–10,000 pg mL−1 | ||||
| miRNA-375 | DPV | miRNA-375 | [109] | ||
| miRNA-141 | 0.01–10 μM | miRNA-141 PSA | |||
| PSA | |||||
| Methotrexate (MTX) | DPV | 35 nM | 5–1000 μM | [121] | |
| Leukemia | Lactate dehydrogenase | 25 U L−1 | 60–700 U L−1 | ||
| Uric acid (UA) | 450 nM | ||||
| Urea | 20 μM | ||||
| Breast Cancer | miRNA-155 | DPV | 0.98 fM | 1 fM–10 nM | [122] |
| miRNA-21 | 3.58 fM | ||||
| miRNA-16 | 0.25 fM | ||||
| miRNA-155 | 0.33 fM | ||||
| miRNA-21 | SWV | 0.04 fM | 0.001–1000 pM | [123] | |
| miRNA-210 | 0.28 fM | ||||
| Rheumatoid Arthritis (RA) | Anti-CCP-ab | EIS | 0.82 IU mL−1 | 1–800 IU mL−1 | [112] |
| CXCL7 | Amperometry | 0.8 ng mL−1 | 1–75 ng mL−1 | [124] | |
| MMP3 | 1.2 pg mL−1 | 2–2000 pg mL−1 | |||
| SLE | BAFF | Amperometry | 0.08 ng mL−1 | 0.24–120 ng mL−1 | [125] |
| Colorectal Cancer | APRIL | 0.06 ng mL−1 | 0.19–25 ng mL−1 |
| Metabolic Syndrome | Biomarker | Optical Method | LOD | Linear Range | Refs. |
|---|---|---|---|---|---|
| Cardiovascular Diseases (CVDs) | Procalcitonin (PCT) | SPR | 1.22 pg mL−1 | 10–105 pg mL−1 | [154] |
| Myoglobin (MG) | SPR | <1 ng mL−1 | 1–25 ng mL−1 | [155] | |
| Cardiac troponin I (cTnI) | <1 ng mL−1 | 1–25 ng mL−1 | |||
| Interleukin-6 (IL-6) | Fiber-optic fluorescence | 5 pM (0.12 ng mL−1) | 5–500 pM | [156] | |
| B-type natriuretic peptide (BNP) Cardiac troponin I (cTnI) C-reactive protein (CRP) Myoglobin (MG) | Fiber-optic fluorescence | 0.1 ng mL−1 7 × 10−3 ng mL−1 700 ng mL−1 70 ng mL−1 | 0.1–1 ng/mL 0.7–7 ng/mL 700–7000 ng/mL 70–700 ng/ml | [157] | |
| Fiber-optic SPR | 1.48 ng mL−1 | 1–1000 ng mL−1 | [158] | ||
| Interleukin-6 (IL-6) | Electrochemical | 0.886 fg mL−1 | 0.1–1000 pg mL−1 | ||
| Prediabetes | Glucose | Fiber-optic SPR | Can be tuned by changing the microgel concentration | 16 μM–16 mM | [159] |
| Glucose | Microfluidics-enabled multi-scattering of light | 110 nM | 1–400 μM | [160] | |
| Lactate | 240 nM | 10–3000 μM | |||
| Glucose | Colorimetric | 27.2 μM | 0.0781–5 mM | [161] | |
| Lactate | 29.6 μM | 0.0391–2.5 mM | |||
| Prostate Cancer | LSPR | 100 fg mL−1 | [162] | ||
| PSA | 50 fgmL−1–5 ngmL−1 | ||||
| PSA | SERS | 0.46 fg mL−1 | 0.46 fg mL−1–478.93 ng mL−1 | [164] | |
| PSMA | 1.05 fg mL−1 | 1.05 fg mL−1–113.4 ng mL−1 | |||
| hK2 | 0.67 fg mL−1 | 0.67 fg mL−1–466.23 ng mL−1 | |||
| PSA | SERS | 0.37 pg mL−1 | 1 pg mL−1–10 µg mL−1 | [165] | |
| CEA | 0.43 pg mL−1 | 10 pg mL−1–1 µg mL−1 | |||
| AFP | 0.26 pg mL−1 | 10 pg mL−1–1 µg mL−1 | |||
| PSA | SERS | 10 pg mL−1 for all proteins | - | [166] | |
| CEA | |||||
| AFP | |||||
| Multiple Cancers | AFP | Silicon photonic sensor array | - | [168] | |
| ALCAM | - | ||||
| CA15-3 | - | ||||
| CA19-9 | |||||
| CA-125 | |||||
| CEA | |||||
| Osteopontin | |||||
| PSA | |||||
| Rheumatoid Arthritis (RA) | miRNA-21 | FRET | 1 pM (both) | 1 pM–1 nM (both) | [169] |
| miRNA-155 | |||||
| miRNA-21 | SPR | 10 aM (both) | 10 aM–10 pM (both) | [170] | |
| miRNA-155 | |||||
| miRNA-21 | Silicon photonic Microring resonators | 9 nM | 20 nM–2 µM | [171] | |
| miRNA-26a | 4 nM | 20 nM–2 µM | |||
| miRNA-29a | <1 nM | 2 nM–2 µM | |||
| miRNA-106a | 2 nM | 2 nM–2 µM | |||
| miRNA-222, miRNA-335 | 1 nM | 2 nM–2 µM | |||
| let-7c-5p | 4 nM | 4–250 nM | |||
| miRNA-21 | Silicon photonic Microring resonators | 4 nM | 4–250 nM | [172] | |
| miRNA-24 | 1.95 nM | 1.95 nM–2 µM | |||
| miRNA-133b | 62.5 nM | 62.5 nM–1 µM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, S.S.; Nguyen, H.A.; Zhang, J.; Tabassum, S.; Cao, H. Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders. Sensors 2022, 22, 5200. https://doi.org/10.3390/s22145200
Chu SS, Nguyen HA, Zhang J, Tabassum S, Cao H. Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders. Sensors. 2022; 22(14):5200. https://doi.org/10.3390/s22145200
Chicago/Turabian StyleChu, Sung Sik, Hung Anh Nguyen, Jimmy Zhang, Shawana Tabassum, and Hung Cao. 2022. "Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders" Sensors 22, no. 14: 5200. https://doi.org/10.3390/s22145200
APA StyleChu, S. S., Nguyen, H. A., Zhang, J., Tabassum, S., & Cao, H. (2022). Towards Multiplexed and Multimodal Biosensor Platforms in Real-Time Monitoring of Metabolic Disorders. Sensors, 22(14), 5200. https://doi.org/10.3390/s22145200







