Image Segmentation and Quantification of Droplet dPCR Based on Thermal Bubble Printing Technology
Abstract
:1. Introduction
2. Materials and Methods
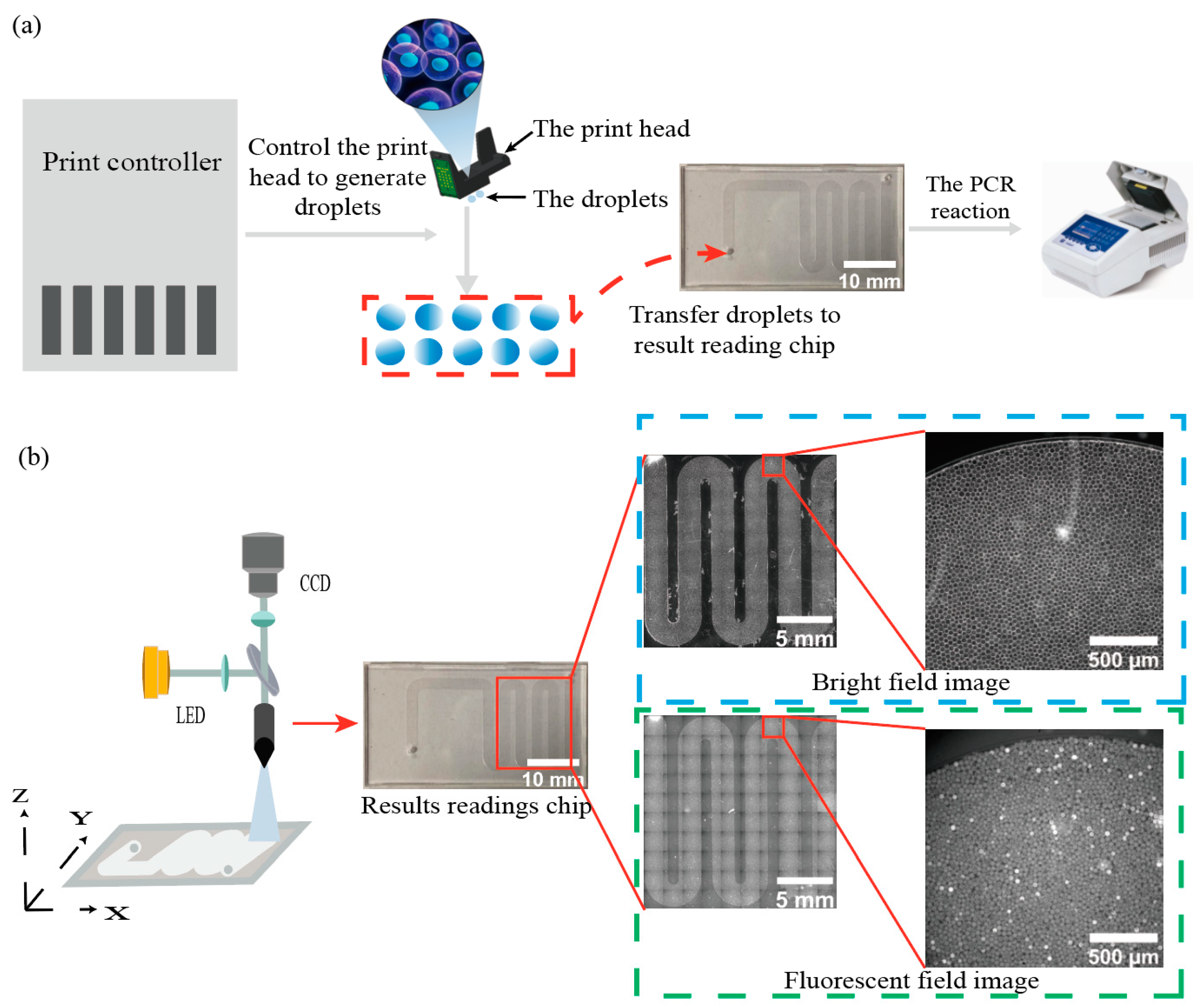
2.1. High-Throughput Droplet Generation and Acquisition of Image Data
2.2. Droplet Identification, Localization, and Noise Filtering
2.3. Droplet Segmentation in the Fluorescent Image and Signal Quantification
- Gaussian filter operator is used to blurring an image of the uniform droplet in the fluorescence channel. The blurred image (Figure 3a) is used as the vignetting correction template. The 3 × 3 Gaussian template g(x, y) is shown in Equation (4) and multiplied by a constant 1/16 to achieve a weighted average of the pixel values in the 3 × 3 neighborhood [40].
- 2.
- A quadratic polynomial is chosen as the fitting formula, and the pixel gray value surface is fitted to the template image using the least-squares method, and the surface fitting function is shown in Equation (5) [41,42]. The vignetting correction template is sampled to find the surface fitting coefficients, and the fitted surface is drawn as shown in Figure 3b. The maximum value of the fitted surface equation is divided by the surface fitting value of the image, and the result is normalized to obtain the normalized dark corner surface compensation model as shown in Figure 3c.
- 3.
- The calculated vignetting compensation model is used to correct the images captured by the camera to eliminate image vignetting. Figure 3d shows the image without vignetting correction, and the gray value of the image is counted along the yellow diagonal line in Figure 3d and the gray value curve is plotted as shown in Figure 3e. The image after vignetting correction and the gray value curve of the image are shown in Figure 3f,g. Comparing Figure 3e,g, it is obvious that the image vignetting is better compensated. The pixel value surfaces of the images before and after vignetting correction are shown in Figure S3a,b. The average values of the difference between the average pixel values of the eight edge regions and the average pixel values of the center region in the attached Figure S4a,b are calculated as shown in the attached Tables S1 and S2, and the results are 667 and 71, respectively, and the uniformity of the images after vignetting correction significantly improved. The satellite droplets affect the accuracy of the fluorescence field droplet signal calculation, and the pixel values of each droplet region are sorted and selected as the signal value of this droplet by averaging the 40–60% length part of the sequence, and Figure S5 represents the segmented droplet signal image after vignetting correction, and the satellite droplets are marked in the orange box.
3. Results
3.1. Bright-Field Image Droplet Segmentation Accuracy
3.2. Fluorescence Image Droplet Classification Accuracy
3.3. Validation of Positive Droplet Concentration Gradient Detection with Multiple Fluorescence Channels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suo, T.; Liu, X.J.; Feng, J.P.; Guo, M.; Hu, W.J.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.H.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef]
- Tan, C.R.; Fan, D.D.; Wang, N.; Wang, F.; Wang, B.; Zhu, L.X.; Guo, Y. Applications of digital PCR in COVID-19 pandemic. View 2021, 2, 20200082. [Google Scholar] [CrossRef]
- Tan, S.Y.H.; Kwek, S.Y.M.; Low, H.; Pang, Y.L.J. Absolute quantification of SARS-CoV-2 with Clarity Plus™ digital PCR. Methods 2022, 201, 26–33. [Google Scholar] [CrossRef]
- Caswell, R.C.; Snowsill, T.; Houghton, J.A.L.; Chakera, A.J.; Shepherd, M.H.; Laver, T.W.; Knight, B.A.; Wright, D.; Hattersley, A.T.; Ellard, S. Noninvasive Fetal Genotyping by Droplet Digital PCR to Identify Maternally Inherited Monogenic Diabetes Variants. Clin. Chem. 2020, 66, 958–965. [Google Scholar] [CrossRef]
- Jacky, L.; Yurk, D.; Alvarado, J.; Leatham, B.; Schwartz, J.; Annaloro, J.; MacDonald, C.; Rajagopal, A. Virtual-Partition Digital PCR for High-Precision Chromosomal Counting Applications. Anal. Chem. 2021, 93, 17020–17029. [Google Scholar] [CrossRef]
- Manderstedt, E.; Nilsson, R.; Ljung, R.; Lind-Hallden, C.; Astermark, J.; Hallden, C. Detection of mosaics in hemophilia A by deep Ion Torrent sequencing and droplet digital PCR. Res. Pract. Thromb. Haemost. 2020, 4, 1121–1130. [Google Scholar] [CrossRef]
- Sawakwongpra, K.; Tangmansakulchai, K.; Ngonsawan, W.; Promwan, S.; Chanchamroen, S.; Quangkananurug, W.; Sriswasdi, S.; Jantarasaengaram, S.; Ponnikorn, S. Droplet-based digital PCR for non-invasive prenatal genetic diagnosis of alpha and beta-thalassemia. Biomed. Rep. 2021, 15, 82. [Google Scholar] [CrossRef]
- Olmedillas-Lopez, S.; Olivera-Salazar, R.; Garcia-Arranz, M.; Garcia-Olmo, D. Current and Emerging Applications of Droplet Digital PCR in Oncology: An Updated Review. Mol. Diagn. Ther. 2022, 26, 61–87. [Google Scholar] [CrossRef]
- Palacin-Aliana, I.; Garcia-Romero, N.; Asensi-Puig, A.; Carrion-Navarro, J.; Gonzalez-Rumayor, V.; Ayuso-Sacido, A. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines 2021, 9, 906. [Google Scholar] [CrossRef]
- Powell, L.; Dhummakupt, A.; Siems, L.; Singh, D.; Le Duff, Y.; Uprety, P.; Jennings, C.; Szewczyk, J.; Chen, Y.; Nastouli, E.; et al. Clinical validation of a quantitative HIV-1 DNA droplet digital PCR assay: Applications for detecting occult HIV-1 infection and monitoring cell-associated HIV-1 dynamics across different subtypes in HIV-1 prevention and cure trials. J. Clin. Virol. 2021, 139, 104822. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Sreejith, K.R.; Ooi, C.H.; Jin, J.; Dao, D.V.; Nguyen, N.-T. Digital polymerase chain reaction technology—Recent advances and future perspectives. Lab Chip 2018, 18, 3717–3732. [Google Scholar] [CrossRef]
- Li, H.T.; Pan, J.Z.; Fang, Q. Development and Application of Digital PCR Technology. Prog. Chem. 2020, 32, 581–593. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Salipante, S.J.; Jerome, K.R. Digital PCR-An Emerging Technology with Broad Applications in Microbiology. Clin. Chem. 2020, 66, 117–123. [Google Scholar] [CrossRef]
- Wang, Y.T.; Zhang, X.X.; Shang, L.R.; Zhao, Y.J. Thriving microfluidic technology. Sci. Bull. 2021, 66, 9–12. [Google Scholar] [CrossRef]
- Heyries, K.A.; Tropini, C.; VanInsberghe, M.; Doolin, C.; Petriv, O.I.; Singhal, A.; Leung, K.; Hughesman, C.B.; Hansen, C.L. Megapixel digital PCR. Nat. Methods 2011, 8, 649–651. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Hatch, A.C.; Fisher, J.S.; Tovar, A.R.; Hsieh, A.T.; Lin, R.; Pentoney, S.L.; Yang, D.L.; Lee, A.P. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 2011, 11, 3838–3845. [Google Scholar] [CrossRef]
- Hu, Z.; Fang, W.; Gou, T.; Wu, W.; Hu, J.; Zhou, S.; Mu, Y. A novel method based on a Mask R-CNN model for processing dPCR images. Anal. Methods 2019, 11, 3410–3418. [Google Scholar] [CrossRef]
- Shen, J.; Zheng, J.; Li, Z.; Liu, Y.; Jing, F.; Wan, X.; Yamaguchi, Y.; Zhuang, S. A rapid nucleic acid concentration measurement system with large field of view for a droplet digital PCR microfluidic chip. Lab Chip 2021, 21, 3742–3747. [Google Scholar] [CrossRef]
- Gale, B.K.; Jafek, A.R.; Lambert, C.J.; Goenner, B.L.; Moghimifam, H.; Nze, U.C.; Kamarapu, S.K. A Review of Current Methods in Microfluidic Device Fabrication and Future Commercialization Prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef]
- Guo, M.; Li, Y.; Su, Y.; Lambert, T.; Nogare, D.D.; Moyle, M.W.; Duncan, L.H.; Ikegami, R.; Santella, A.; Rey-Suarez, I.; et al. Rapid image deconvolution and multiview fusion for optical microscopy. Nat. Biotechnol. 2020, 38, 1337–1346. [Google Scholar] [CrossRef]
- Sarder, P.; Nehorai, A. Deconvolution methods for 3-D fluorescence microscopy images. IEEE Signal Process. Mag. 2006, 23, 32–45. [Google Scholar] [CrossRef]
- Sinaga, K.P.; Yang, M. Unsupervised K-Means Clustering Algorithm. IEEE Access 2020, 8, 80716–80727. [Google Scholar] [CrossRef]
- Bian, X.; Jing, F.; Li, G.; Fan, X.; Jia, C.; Zhou, H.; Jin, Q.; Zhao, J. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes. Biosens. Bioelectron. 2015, 74, 770–777. [Google Scholar] [CrossRef]
- Bu, W.; Li, W.; Li, J.; Ao, T.; Li, Z.; Wu, B.; Wu, S.; Kong, W.; Pan, T.; Ding, Y.; et al. A low-cost, programmable, and multi-functional droplet printing system for low copy number SARS-CoV-2 digital PCR determination. Sens. Actuators B-Chem. 2021, 348, 130678. [Google Scholar] [CrossRef]
- Meng, X.; Yu, Y.; Jin, G. Numerical Simulation and Experimental Verification of Droplet Generation in Microfluidic Digital PCR Chip. Micromachines 2021, 12, 409. [Google Scholar] [CrossRef]
- Pan, Y.; Ma, T.; Meng, Q.; Mao, Y.; Chu, K.; Men, Y.; Pan, T.; Li, B.; Chu, J. Droplet digital PCR enabled by microfluidic impact printing for absolute gene quantification. Talanta 2020, 211, 120680. [Google Scholar] [CrossRef]
- OpenCV SimpleBlobDetector. Available online: https://learnopencv.com/blob-detection-using-opencv-python-c/ (accessed on 10 August 2022).
- Dejgaard, S.Y.; Presley, J.F. New Automated Single-Cell Technique for Segmentation and Quantitation of Lipid Droplets. J. Histochem. Cytochem. 2014, 62, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.Y.; Lee, S.S.; Hsu, Y.H. Development of an imaging method for quantifying a large digital PCR droplet. In Proceedings of the Conference on Optical Diagnostics and Sensing XVII—Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 30–31 January 2017. [Google Scholar] [CrossRef]
- Sanka, I.; Bartkova, S.; Pata, P.; Smolander, O.-P.; Scheler, O. Investigation of Different Free Image Analysis Software for High-Throughput Droplet Detection. ACS Omega 2021, 6, 22625–22634. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, S.; Donovan, J.; Macpherson, R.; Tormey, D. In-process analysis of pharmaceutical emulsions using computer vision and artificial intelligence. Chem. Eng. Res. Des. 2021, 166, 281–294. [Google Scholar] [CrossRef]
- Anees, V.M.; Kumar, G.S. Direction estimation of crowd flow in surveillance videos. In Proceedings of the 2017 IEEE Region 10 Symposium (TENSYMP), Cochin, India, 14–16 July 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Mishra, R.K.; Jain, P. A system on chip based serial number identification using computer vision. In Proceedings of the 2016 IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT), Bangalore, India, 20–21 May 2016; pp. 278–283. [Google Scholar] [CrossRef]
- Likar, B.; Maintz, J.B.A.; Viergever, M.A.; Pernus, F. Retrospective shading correction based on entropy minimization. J. Microsc. 2000, 197, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-C.; Zheng, X.-F.; Mao, X.-Y.; Cheng, H.-B.; Chen, Y. Vignetting compensation in the collection process of LED display camera. Chin. J. Liq. Cryst. Disp. 2019, 34, 778–786. [Google Scholar] [CrossRef]
- Zheng, Y.; Lin, S.; Kambhamettu, C.; Yu, J.; Kang, S.B. Single-Image Vignetting Correction. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 2243–2256. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Li, X.M.; Zhang, C.M.; Yue, Y.Z.; Wang, K.P. Cubic surface fitting to image by combination. Sci. China-Inf. Sci. 2010, 53, 1287–1295. [Google Scholar] [CrossRef] [Green Version]
- Mieloch, K.; Mihailescu, P.; Munk, A. Dynamic threshold using polynomial surface regression with application to the binarisation of fingerprints. In Proceedings of the Conference on Biometric Technology for Human Identification II, Orlando, FL, USA, 28–29 March 2005; pp. 94–104. [Google Scholar]
- Abualigah, L.M.; Khader, A.T. Unsupervised text feature selection technique based on hybrid particle swarm optimization algorithm with genetic operators for the text clustering. J. Supercomput. 2017, 73, 4773–4795. [Google Scholar] [CrossRef]





| Number of Bright Field Images | Total Number of Droplets | Number of Droplet Segmentation | Number of Droplets Actually Segmented | Number of Segmentation Errors | Segmentation Accuracy (%) |
| Workflow 1 | |||||
| 1 | 1917 | 1859 | 1843 | 16 | 96.14% |
| 5 | 8092 | 7724 | 7612 | 112 | 94.07% |
| 10 | 17,165 | 16,102 | 15,797 | 305 | 92.03% |
| 20 | 33,136 | 29,100 | 28,223 | 877 | 85.17% |
| Workflow 2 | |||||
| 1 | 1917 | 1771 | 1764 | 7 | 92.02% |
| 5 | 8092 | 7214 | 7150 | 64 | 88.36% |
| 10 | 17,165 | 14,869 | 14,733 | 136 | 85.83% |
| 20 | 33,136 | 27,506 | 27,233 | 273 | 82.19% |
| Worflow 3 | |||||
| 1 | 1917 | 1725 | 1721 | 4 | 89.78% |
| 5 | 8092 | 6998 | 6977 | 21 | 86.22% |
| 10 | 17,165 | 14,303 | 14,264 | 39 | 83.10% |
| 20 | 33,136 | 25,573 | 25,476 | 97 | 76.88% |
| Workflow with SimpleBlobDetector algorithm | |||||
| 1 | 1917 | 1907 | 1905 | 2 | 99.37% |
| 5 | 8092 | 8052 | 8037 | 15 | 99.32% |
| 10 | 17,165 | 17,075 | 17,047 | 28 | 99.31% |
| 20 | 33,136 | 33,004 | 32,954 | 50 | 99.45% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Shan, Z.; Ning, W.; Wu, X. Image Segmentation and Quantification of Droplet dPCR Based on Thermal Bubble Printing Technology. Sensors 2022, 22, 7222. https://doi.org/10.3390/s22197222
Zhu M, Shan Z, Ning W, Wu X. Image Segmentation and Quantification of Droplet dPCR Based on Thermal Bubble Printing Technology. Sensors. 2022; 22(19):7222. https://doi.org/10.3390/s22197222
Chicago/Turabian StyleZhu, Mingjie, Zilong Shan, Wei Ning, and Xuanye Wu. 2022. "Image Segmentation and Quantification of Droplet dPCR Based on Thermal Bubble Printing Technology" Sensors 22, no. 19: 7222. https://doi.org/10.3390/s22197222




