A Parallel Cross Convolutional Recurrent Neural Network for Automatic Imbalanced ECG Arrhythmia Detection with Continuous Wavelet Transform
Abstract
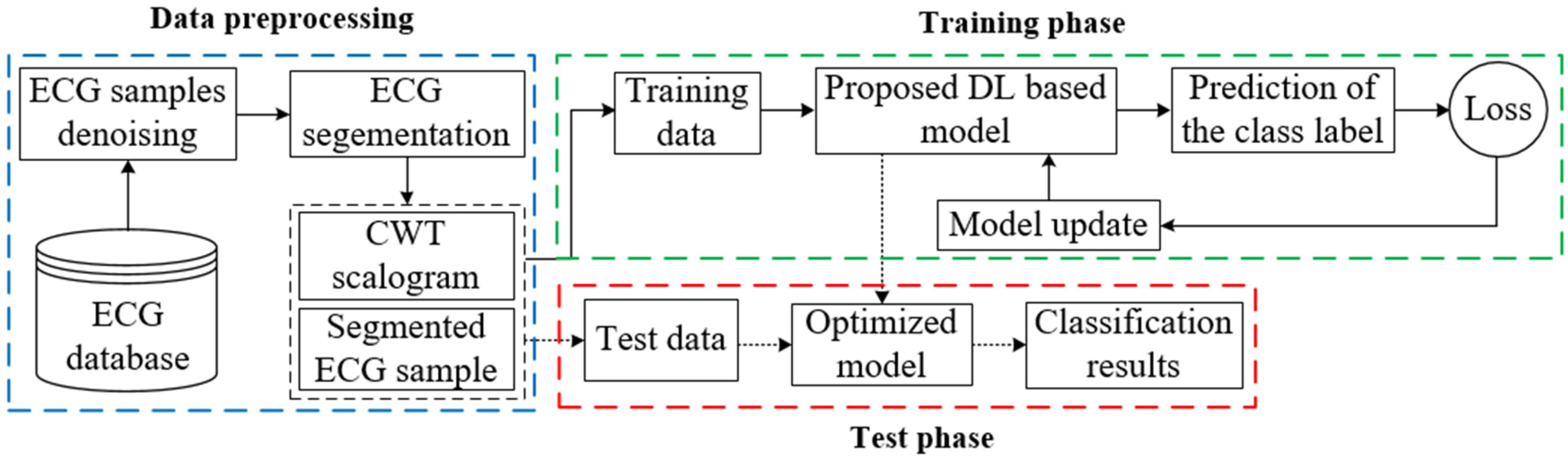
:1. Introduction
- ▪
- Considering ECG arrhythmia detection as a multi-class imbalance problem, this paper proposes a novel parallel cross convolutional RNN classifier to improve detection performance.
- ▪
- The proposed model includes two branches consisting of the LSTM network and CNN to capture both temporal characteristics from the ECG segment and spatial characteristics from the 2D scalogram. The network becomes more distinguishable and robust through cross-fusion of the features.
- ▪
- Continuous wavelet transform (CWT) has been adopted to convert the preprocessed ECG segments into 2D scalograms. Subsequently, both ECG segment and corresponding 2D scalogram have been used as input for training and testing the proposed model.
- ▪
- Finally, we have evaluated the performances of the proposed model on the MIT-BIH arrhythmia dataset. We also compared the performance with other existing works and investigated the performance by varying the wavelet function, such as Gaussian wavelets of order 8 (gaus8) and order 4 (gaus4), Mexican hat wavelet (mexh), Morlet wavelet (morl).
2. Methodology
2.1. ECG Signal Preprocessing and Segmentation
2.2. Scalogram Obtained Using CWT
2.3. Model Description
3. Experimental Results
3.1. ECG Dataset Description
3.2. Implementation Details and Performance Metrics
3.3. Performance Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 17 May 2017).
- Ahmad, Z.; Tabassum, A.; Guan, L.; Khan, N.M. ECG Heartbeat Classification Using Multimodal Fusion. IEEE Access 2021, 9, 100615–100626. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, D.; Liu, A.; Gao, M.; Chen, X.; Zhang, X.; Chen, X. MLBF-Net: A Multi-Lead-Branch Fusion Network for Multi-Class Arrhythmia Classification Using 12-Lead ECG. IEEE J. Transl. Eng. Health Med. 2021, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Chen, X.; Wang, Y.; Wang, L. Arrhythmia Recognition and Classification Using ECG Morphology and Segment Feature Analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Osowski, S.; Hoa, L.T.; Markiewic, T. Support vector machine based expert system for reliable heartbeat recognition. IEEE Trans. Biomed. Eng. 2004, 51, 582–589. [Google Scholar] [CrossRef]
- Chazal, P.D.; O’Dwyer, M.; Reilly, R.B. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar]
- Jun, T.J.; Park, H.J.; Kim, Y.H. Premature ventricular contraction beat detection with deep neural networks. In Proceedings of the 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA), Anaheim, CA, USA, 18–20 December 2016; pp. 859–864. [Google Scholar]
- Monasterio, V.; Laguna, P.; Martinez, J.P. Multilead analysis of t-wave alternans in the ecg using principal component analysis. IEEE Trans. Biomed. Eng. 2009, 56, 1880–1890. [Google Scholar] [CrossRef]
- Özbay, Y.; Tezel, G. A new method for classification of ECG arrhythmias using neural network with adaptive activation function. Digit. Signal Process. 2010, 20, 1040–1049. [Google Scholar] [CrossRef]
- Tuncer, T.; Dogan, S.; Pławiak, P.; Acharya, U.R. Automated arrhythmia detection using novel hexadecimal local pattern and multilevel wavelet transform with ECG signals. Knowl. Based Syst. 2019, 186, 104923. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Luo, X.; Choi, K.S.; Wu, X. Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 2014, 46, 79–89. [Google Scholar] [CrossRef]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 2012, 59, 2930–2941. [Google Scholar]
- Chen, S.; Hua, W.; Li, Z.; Li, J.; Gao, X. Heartbeat classification using projected and dynamic features of ECG signal. Biomed. Signal Process. Control 2017, 31, 165–173. [Google Scholar] [CrossRef]
- Jian, L.; Shuang, S.; Guozhong, S.; Yu, F. Classification of ECG Arrhythmia Using CNN, SVM and LDA. In International Conference on Artificial Intelligence and Security; Lecture Notes in Computer Science (LNCS 11633); Springer: Berlin/Heidelberg, Germany, 2019; pp. 191–201. [Google Scholar]
- Nurmaini, S.; Tondas, A.E.; Darmawahyuni, A.; Rachmatullah, M.N.; Khoirani, R. Robust detection of atrial fibrillation from short-term electrocardiogram using convolutional neural networks. Future Gener. Comput. Syst. 2020, 113, 304–317. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf. Sci. 2017, 405, 81–90. [Google Scholar] [CrossRef]
- Wang, H.; Shi, H.; Lin, K.; Qin, C.; Zhao, L.; Huang, Y.; Liu, C. A high precision arrhythmia classification method based on dual fully connected neural network. Biomed. Signal Process. Control 2020, 58, 101874. [Google Scholar] [CrossRef]
- Chen, A.; Wang, F.; Liu, W.; Chang, S.; Wang, H.; He, J.; Huang, Q. Multi-information fusion neural networks for arrhythmia automatic detection. Comput. Meth. Programs Biomed. 2020, 193, 105479. [Google Scholar] [CrossRef]
- Chen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2020, 57, 101819. [Google Scholar] [CrossRef]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG arrhythmia classification using STFT-based spectrogram and convolutional neural network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Hao, C.; Wibowo, S.; Majmudar, M.; Rajput, K.S. Spectro-temporal feature based multi-channel convolutional neural network for ECG beat classification. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 5642–5645. [Google Scholar]
- Oliveira, A.T.; Nobrega, E.G.O. A novel arrhythmia classification method based on convolutional neural networks interpretation of electrocardiogram images. In Proceedings of the 2019 IEEE International Conference on Industrial Technology (ICIT), Melbourne, VIC, Australia, 13–15 February 2019; pp. 841–846. [Google Scholar]
- Al Rahhal, M.M.; Bazi, Y.; Almubarak, H.; Alajlan, N.; Al Zuair, M. Dense convolutional networks with focal loss and image generation for electrocardiogram classification. IEEE Access 2019, 7, 182225–182237. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, M.; Zhang, Y.; Liao, Y.; Huang, Q.; Chang, S.; Wang, H.; He, J. Real-time multilead convolutional neural network for myocardial infarction detection. IEEE J. Biomed. Health Inform. 2018, 22, 1434–1444. [Google Scholar] [CrossRef]
- Wang, T.; Lu, C.; Sun, Y.; Yang, M.; Liu, C.; Ou, C. Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy 2021, 23, 119. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Jiang, J.; Zhang, H.; Pi, D.; Dai, C. A novel multi-module neural network system for imbalanced heartbeats classification. Expert Syst. Appl. X 2019, 1, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Wei, S.; Zhou, F.; Li, D. Heartbeats Classification Using Hybrid Time-Frequency Analysis and Transfer Learning Based on ResNet. IEEE J. Biomed. Health Inform. 2021, 25, 4175–4184. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Guo, D.; Song, M.; Teng, Y.; Wang, X. PCCN: Parallel Cross Convolutional Neural Network for Abnormal Network Traffic Flows Detection in Multi-Class Imbalanced Network Traffic Flows. IEEE Access 2019, 7, 119904–119916. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Lee, H.K.; Choi, Y.-S. Application of Continuous Wavelet Transform and Convolutional Neural Network in Decoding Motor Imagery Brain-Computer Interface. Entropy 2019, 21, 1199. [Google Scholar] [CrossRef]
- Guo, M.F.; Zeng, X.D.; Chen, D.Y.; Yang, N.C. Deep-learning-based earth fault detection using continuous wavelet transform and convolutional neural network in resonant grounding distribution systems. IEEE Sens. J. 2017, 18, 1291–1300. [Google Scholar] [CrossRef]
- Wang, L.; Wang, C.; Fu, F.; Yu, X.; Guo, H.; Xu, C.; Jing, X.; Zhang, H.; Dong, X. Temporal Lobe seizure prediction based on a complex Gaussian wavelet. Clin. Neurophysiol. 2011, 4, 656–663. [Google Scholar] [CrossRef]
- Auger, F.; Patrick, F.; Paulo, G.; Olivier, L. Time-Frequency Toolbox; CNRS France-Rice University: Paris, France, 1996. [Google Scholar]
- Meignen, S.; Oberlin, T.; McLaughlin, S. A New Algorithm for Multicomponent Signals Analysis Based on Synchro Squeezing: With an Application to Signal Sampling and Denoising. IEEE Trans. Signal Process. 2012, 60, 5787–5798. [Google Scholar] [CrossRef]
- Kong, W.; Dong, Z.Y.; Hill, D.J.; Luo, F.; Xu, Y. Short-term residential load forecasting based on resident behavior learning. IEEE Trans. Power Syst. 2018, 33, 1087–1088. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Mar, T.; Zaunseder, S.; Martínez, J.P.; Llamedo, M.; Poll, R. Optimization of ECG classification by means of feature selection. IEEE Trans. Biomed. Eng. 2011, 58, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Mondéjar-Guerra, V.; Novo, J.; Rouco, J.; Penedo, M.G.; Ortega, M. Heartbeat classification fusing temporal and morphological information of ECGs via ensemble of classifiers. Biomed. Signal Process. Control 2019, 47, 41–48. [Google Scholar] [CrossRef]









| Top Branch | Bottom Branch | ||
|---|---|---|---|
| Layer | Output Volume | Layer | Output Volume |
| Input_1 | (200, 1) | Input_2 | (100, 100, 1) |
| Lstm_unit_1 | (200, 16) | Conv_block_1 | (50, 50, 16) |
| Reshape_1 | (4, 50, 16) | - | - |
| Concatenate_1 | (54, 50, 16) | ||
| Reshape_2 | (2700, 16) | - | - |
| Lstm_unit_2 | (2700, 32) | Conv_block_2 | (27, 25, 32) |
| Reshape_3 | (108, 25, 32) | - | - |
| Concatenate_2 | (135, 25, 32) | ||
| Reshape_4 | (3375, 32) | - | - |
| Lstm_unit_3 | (3375, 64) | Conv_block_3 | (68, 13, 64) |
| Flatten_1 | (216000) | Flatten_2 | (56576) |
| Concatenate_3 | (272576) | ||
| Dropout | (272576) | ||
| Dense_1 | (128) | ||
| Dense_2 | (4) | ||
| AAMI Classes | Class Symbol | Heartbeat Types | Training Samples | Test Samples |
|---|---|---|---|---|
| Normal Beat | NB | Normal beat (NOR) | 72,000 | 18,000 |
| Right bundle branch block beat (RBBB) | ||||
| Left bundle branch block beat (LBBB) | ||||
| Atrial escape beat (AE) | ||||
| Nodal (junctional) escape beat (NE) | ||||
| Supraventricular Ectopic Beat | SVEB | Atrial premature beat (AP) | 2160 | 540 |
| Premature or ectopic supraventricular beat (SP) | ||||
| Nodal premature beat (NP) | ||||
| Aberrated atrial premature beat (AAP) | ||||
| Ventricular Ectopic Beat | VEB | Ventricular escape beat (VE) | 5600 | 1400 |
| Premature ventricular contraction (PVC) | ||||
| Fusion Beat | FB | Fusion of ventricular and normal beat (FVN) | 641 | 160 |
| Hyper-Parameter | Values |
|---|---|
| Training data shape | (80401, 100, 100, 1) and (80401, 200, 1) |
| Test data shape | (20100, 100, 100, 1) and (20100, 200, 1) |
| Batch size | 512 |
| Learning rate | 0.0005, 0.0001, 0.00005 |
| Epoch | 60 |
| Optimizer | Adam |
| Loss function | Categorical-cross entropy |
| Classes | Metrics | lr = 0.0005 | lr = 0.0001 | lr = 0.00005 |
|---|---|---|---|---|
| NB | ACC | 99.25 | 99.48 | 99.37 |
| PPV | 99.41 | 99.62 | 99.35 | |
| SE | 99.75 | 99.79 | 99.94 | |
| F1 | 99.58 | 99.71 | 99.64 | |
| SVEB | ACC | 99.52 | 99.67 | 99.63 |
| PPV | 95.49 | 93.56 | 98.34 | |
| SE | 86.29 | 94.07 | 87.77 | |
| F1 | 90.67 | 93.81 | 92.75 | |
| VEB | ACC | 99.79 | 99.84 | 99.80 |
| PPV | 99.13 | 99.49 | 99.49 | |
| SE | 97.79 | 98.28 | 97.71 | |
| F1 | 98.45 | 98.89 | 98.59 | |
| FB | ACC | 99.74 | 99.83 | 99.76 |
| PPV | 81.29 | 93.71 | 91.17 | |
| SE | 86.88 | 83.75 | 77.50 | |
| F1 | 83.99 | 88.45 | 83.78 | |
| Average | ACC | 99.58 | 99.71 | 99.64 |
| PPV | 93.83 | 96.60 | 97.09 | |
| SE | 92.68 | 93.97 | 90.73 | |
| F1 | 93.17 | 95.22 | 93.69 |
| K-Folds | NB | SVEB | VEB | FB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPV | SE | F1 | PPV | SE | F1 | PPV | SE | F1 | PPV | SE | F1 | |
| Fold = 1 | 99.41 | 99.94 | 99.68 | 96.80 | 89.63 | 93.08 | 99.27 | 97.00 | 98.12 | 97.10 | 82.72 | 89.33 |
| Fold = 2 | 99.51 | 99.80 | 99.66 | 97.54 | 88.15 | 92.61 | 98.14 | 98.00 | 98.07 | 85.19 | 86.25 | 85.71 |
| Fold = 3 | 99.67 | 99.88 | 99.77 | 96.98 | 95.19 | 96.07 | 99.42 | 97.57 | 98.49 | 88.61 | 87.50 | 88.05 |
| Fold = 4 | 99.33 | 99.92 | 99.62 | 98.73 | 86.67 | 92.31 | 98.83 | 96.43 | 97.61 | 78.95 | 75.00 | 76.92 |
| Fold = 5 | 99.22 | 99.88 | 99.55 | 96.76 | 88.52 | 92.46 | 98.67 | 95.57 | 97.10 | 89.23 | 72.50 | 80.00 |
| Fold = 6 | 99.40 | 99.81 | 99.61 | 99.40 | 99.81 | 99.61 | 98.70 | 97.57 | 98.13 | 88.46 | 85.19 | 86.79 |
| Fold = 7 | 99.57 | 99.82 | 99.69 | 94.25 | 91.11 | 92.66 | 98.86 | 99.00 | 98.93 | 98.46 | 80.00 | 88.28 |
| Fold = 8 | 99.60 | 99.78 | 99.69 | 94.74 | 93.68 | 94.21 | 97.58 | 98.00 | 97.79 | 95.31 | 76.25 | 84.72 |
| Fold = 9 | 99.31 | 99.80 | 99.56 | 92.02 | 89.30 | 90.64 | 99.12 | 96.71 | 97.90 | 91.80 | 70.00 | 79.43 |
| Fold = 10 | 99.57 | 99.78 | 99.67 | 93.04 | 94.07 | 93.55 | 98.56 | 97.86 | 98.21 | 93.65 | 73.75 | 82.52 |
| Average | 99.46 | 99.84 | 99.65 | 96.03 | 91.61 | 93.72 | 98.72 | 97.37 | 98.04 | 90.68 | 78.92 | 84.18 |
| Classes | Metrics | Other Studies | Ours | |||||
|---|---|---|---|---|---|---|---|---|
| Liu et al. [14] | Zhang et al. [11] | Ye et al. [12] | Wang et al. [26] | Jiang et al. [28] | Y. Zhang et al. [29] | |||
| NB | PPV | 96.66% | 98.98% | 97.55% | 98.17% | 98.39% | 99.36% | 99.62% |
| SE | 94.06% | 88.94% | 88.61% | 99.42% | 97.64% | 99.62% | 99.79% | |
| F1 | 95.34% | 93.69% | 92.87% | 98.79% | 98.01% | 99.48% | 99.71% | |
| SVEB | PPV | 39.87% | 35.98% | 52.34% | 89.54% | 63.70% | 97.48% | 93.56% |
| SE | 33.12% | 79.06% | 61.02% | 74.56% | 64.40% | 98.44% | 94.07% | |
| F1 | 36.18% | 49.46% | 56.34% | 81.37% | 64.04% | 97.96% | 93.81% | |
| VEB | PPV | 76.51% | 92.75% | 61.45% | 93.25% | 90.00% | 92.66% | 99.49% |
| SE | 90.20% | 85.48% | 81.82% | 95.65% | 91.00% | 87.27% | 98.28% | |
| F1 | 82.79% | 88.96% | 70.19% | 94.43% | 90.50% | 89.88% | 98.89% | |
| FB | PPV | 12.99% | 13.73% | 2.50% | 2.04% | 43.99% | 91.04% | 93.71% |
| SE | 40.72% | 93.81% | 19.69% | 0.26% | 76.70% | 76.25% | 83.75% | |
| F1 | 19.70% | 23.96% | 4.43% | 0.46% | 55.91% | 82.99% | 88.45% | |
| Average | PPV | 56.51% | 60.36% | 53.46% | 70.75% | 74.02% | 95.14% | 96.60% |
| SE | 64.53% | 86.82% | 62.79% | 67.47% | 82.44% | 90.40% | 93.97% | |
| F1 | 58.50% | 64.02% | 55.96% | 68.76% | 77.12% | 92.58% | 95.22% | |
| Classes | Wavelet Functions | ACC | PPV | SE | F1 |
|---|---|---|---|---|---|
| NB | gaus8 | 99.48 | 99.62 | 99.79 | 99.70 |
| gaus4 | 99.38 | 99.38 | 99.93 | 99.66 | |
| mexh | 99.50 | 99.53 | 99.91 | 99.72 | |
| morl | 99.52 | 99.63 | 99.84 | 99.73 | |
| SVEB | gaus8 | 99.67 | 93.55 | 94.07 | 93.81 |
| gaus4 | 99.63 | 98.74 | 87.40 | 92.73 | |
| mexh | 99.72 | 98.01 | 91.67 | 94.74 | |
| morl | 99.71 | 96.35 | 92.96 | 94.62 | |
| VEB | gaus8 | 99.84 | 99.49 | 98.28 | 98.88 |
| gaus4 | 99.82 | 98.92 | 98.50 | 98.71 | |
| mexh | 99.81 | 99.78 | 98.50 | 98.64 | |
| morl | 99.82 | 98.50 | 98.92 | 98.71 | |
| FB | gaus8 | 99.82 | 93.70 | 83.75 | 88.44 |
| gaus4 | 99.79 | 95.41 | 78.13 | 85.91 | |
| mexh | 99.81 | 95.55 | 80.62 | 87.48 | |
| morl | 99.77 | 94.53 | 75.62 | 84.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toma, T.I.; Choi, S. A Parallel Cross Convolutional Recurrent Neural Network for Automatic Imbalanced ECG Arrhythmia Detection with Continuous Wavelet Transform. Sensors 2022, 22, 7396. https://doi.org/10.3390/s22197396
Toma TI, Choi S. A Parallel Cross Convolutional Recurrent Neural Network for Automatic Imbalanced ECG Arrhythmia Detection with Continuous Wavelet Transform. Sensors. 2022; 22(19):7396. https://doi.org/10.3390/s22197396
Chicago/Turabian StyleToma, Tabassum Islam, and Sunwoong Choi. 2022. "A Parallel Cross Convolutional Recurrent Neural Network for Automatic Imbalanced ECG Arrhythmia Detection with Continuous Wavelet Transform" Sensors 22, no. 19: 7396. https://doi.org/10.3390/s22197396





