Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Apparatus and Instruments
2.3. Preparation of AuNPs/CB Modified PGE
2.4. Preparation of Self-Assembled CysAm Monolayer and ssDNA Immobilization
2.5. MicroRNA-21 Hybridization and Electrochemical Measurements
3. Results and Discussion
3.1. Electrochemical Characterization of Different Modified Electrodes
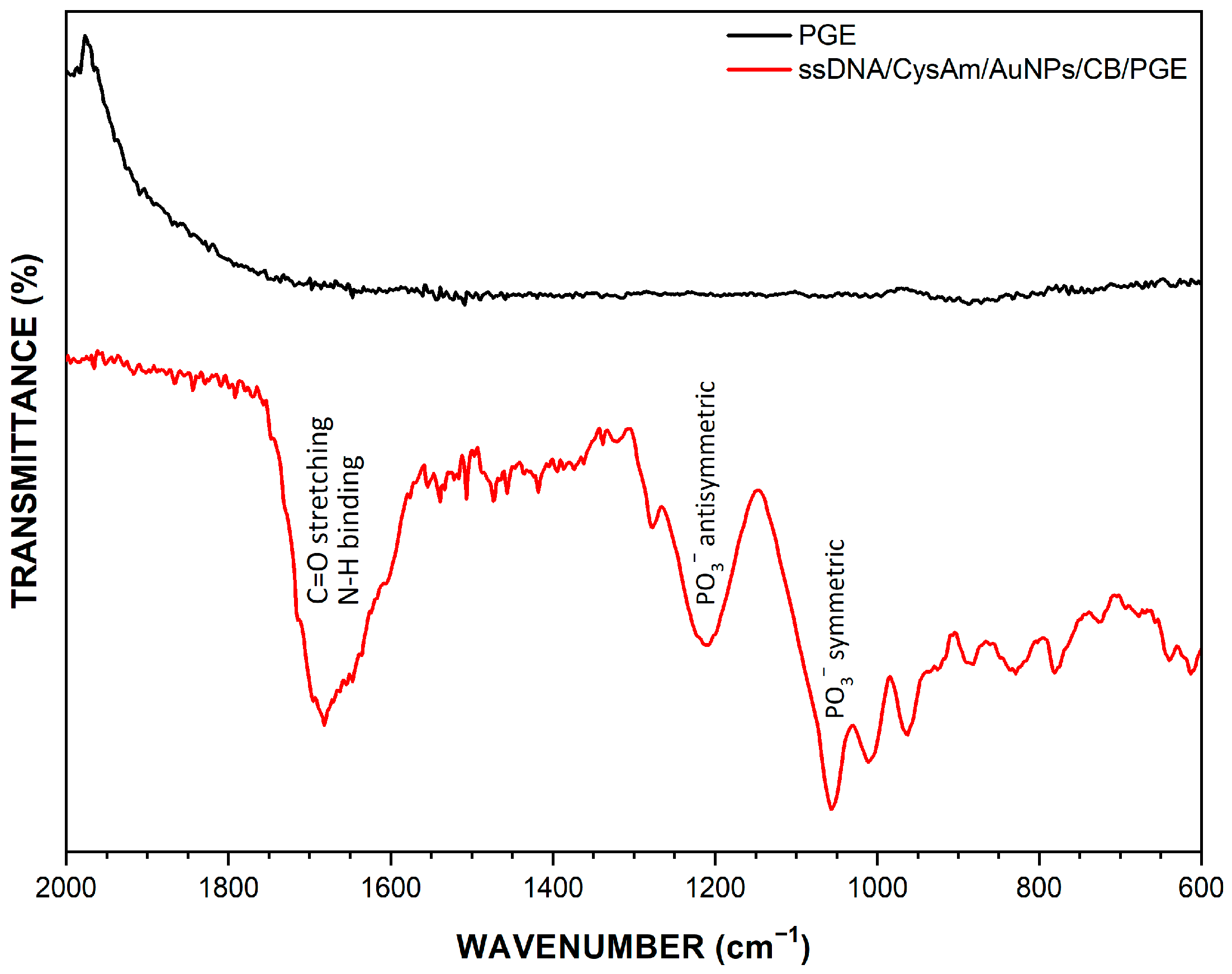
3.2. Characterization of ssDNA Immobilization and Microrna-21 Hybridization
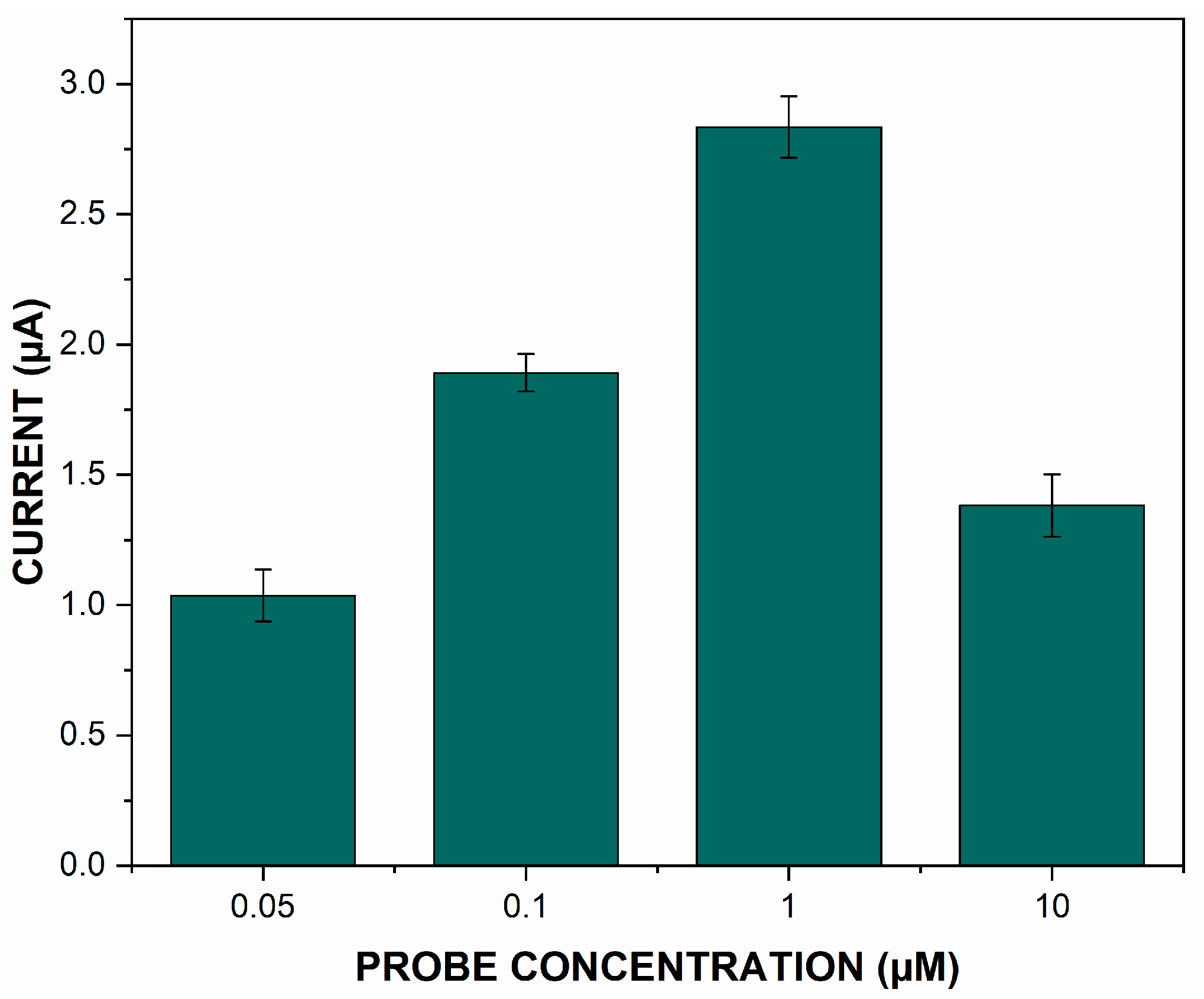
3.3. Optimization of the Immobilized ssDNA Capture Probe Concentration
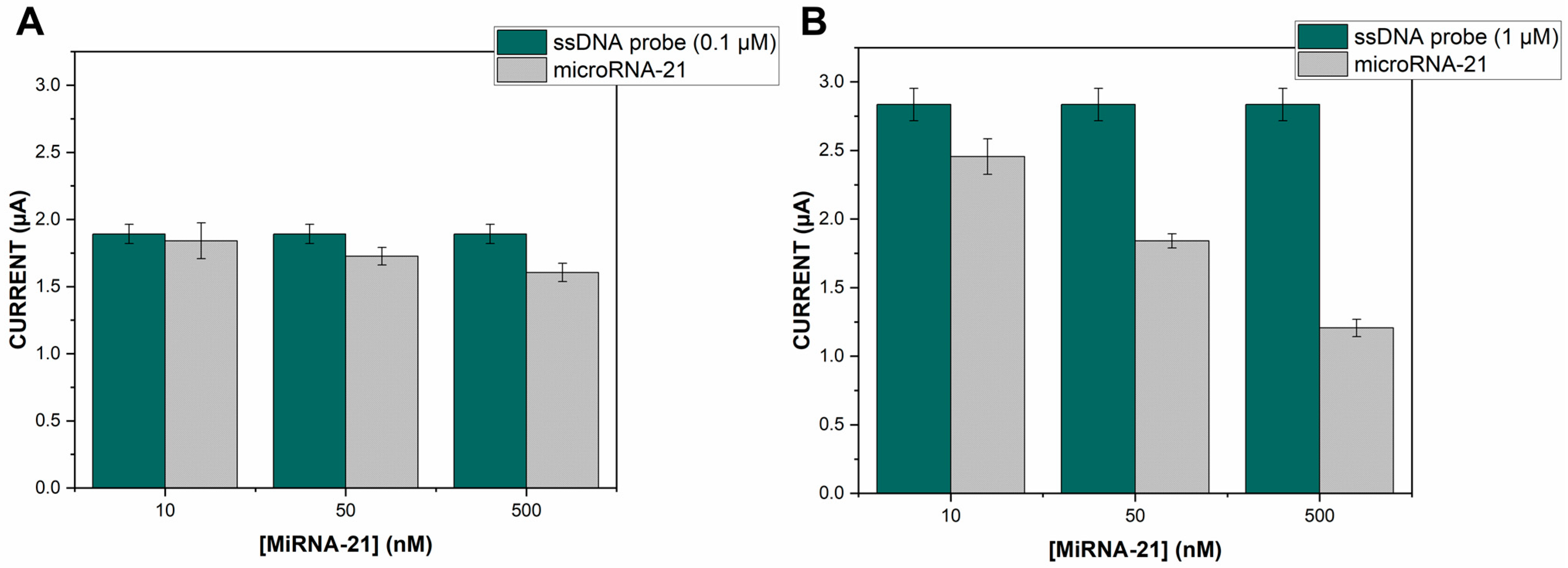
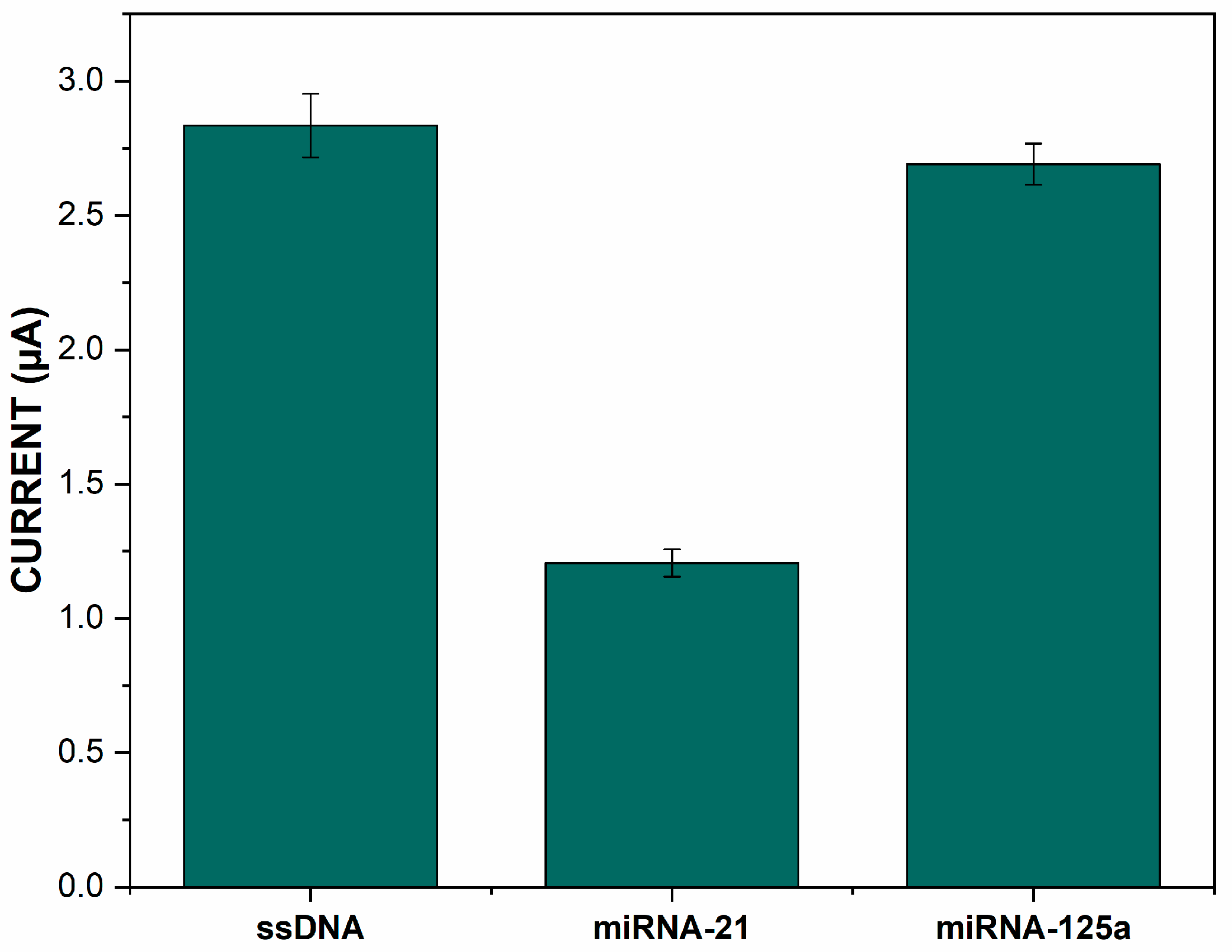
3.4. Hybridization Selectivity and the Quantitative Analysis
3.5. Analysis of microRNA-21 in Serum Sample
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kanzi, A.M.; San, J.E.; Chimukangara, B.; Wilkinson, E.; Fish, M.; Ramsuran, V.; De Oliveira, T. Next generation sequencing and bioinformatics analysis of family genetic inheritance. Front. Genet. 2020, 11, 1250. [Google Scholar] [CrossRef] [PubMed]
- Maljkovic Berry, I.; Melendrez, M.C.; Bishop-Lilly, K.A.; Rutvisuttinunt, W.; Pollett, S.; Talundzic, E.; Morton, L.; Jarman, R.G. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: Approaches, applications, and considerations for development of laboratory capacity. J. Infect. Dis. 2019, 221 (Suppl. S3), S292–S307. Available online: https://academic.oup.com/jid/article/221/Supplement_3/S292/5586940 (accessed on 12 October 2022). [CrossRef] [PubMed] [Green Version]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.-H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2017, 18, 558–576. [Google Scholar] [CrossRef] [Green Version]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Si, H.; Sun, X.; Chen, Y.; Cao, Y.; Chen, S.; Wang, H.; Hu, C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Çakmak, H.A.; Demir, M. MicroRNA and cardiovascular diseases. Balk. Med. J. 2020, 37, 60. [Google Scholar] [CrossRef]
- Shen, M.; Zhou, Y.; Ye, J.; Abdullah AL-maskri, A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, J.; Leger, A.; Prawer, Y.D.J.; A Lane, T.; Harrison, P.J.; Haerty, W.; Clark, M.B. Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 2022, 50, e19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J. High-throughput DNA sequencing-concepts and limitations. BioEssays 2010, 32, 524–536. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Aamri, M.E.; Mohammadi, H.; Amine, A. Novel label-free colorimetric and electrochemical detection for MiRNA-21 based on the complexation of molybdate with phosphate. Microchem. J. 2022, 182, 107851. [Google Scholar] [CrossRef]
- Yammouri, G.; Mandli, J.; Mohammadi, H.; Amine, A. Development of an electrochemical label-free biosensor for microRNA-125a detection using pencil graphite electrode modified with different carbon nanomaterials. J. Electroanal. Chem. 2017, 806, 75–81. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, X.; Liu, P.; Yu, D.-G.; Ge, R. Electrospun nanofiber-based glucose sensors for glucose detection. Front. Chem. 2022, 10, 883. [Google Scholar] [CrossRef]
- Kang, S.; Zhao, K.; Yu, D.-G.; Zheng, X.; Huang, C. Advances in Biosensing and Environmental Monitoring Based on Electrospun Nanofibers. Adv. Fiber Mater. 2022, 4, 404–435. [Google Scholar] [CrossRef]
- Rauf, S.; Lahcen, A.A.; Aljedaibi, A.; Beduk, T.; Filho, J.I.D.O.; Salama, K.N. Gold nanostructured laser-scribed graphene: A new electrochemical biosensing platform for potential point-of-care testing of disease biomarkers. Biosens. Bioelectron. 2021, 180, 113116. [Google Scholar] [CrossRef]
- Annu Sharma, S.; Jain, R.; Raja, A.N. Review—Pencil graphite electrode: An emerging sensing material. J. Electrochem. Soc. 2020, 167, 037501. [Google Scholar] [CrossRef]
- Akanda, M.R.; Sohail, M.; Aziz, M.A.; Kawde, A.N. Recent advances in nanomaterial-modified pencil graphite electrodes for electroanalysis. Electroanalysis 2016, 28, 408–424. [Google Scholar] [CrossRef]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent biomedical applications of gold nanoparticles: A review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Dang, W.; Sun, Y.; Jiao, H.; Xu, L.; Lin, M. AuNPs-NH2/Cu-MOF modified glassy carbon electrode as enzyme-free electrochemical sensor detecting H2O2. J. Electroanal. Chem. 2020, 856, 113592. [Google Scholar] [CrossRef]
- Yang, X.; Feng, M.; Xia, J.; Zhang, F.; Wang, Z. An electrochemical biosensor based on AuNPs/Ti3C2 MXene three-dimensional nanocomposite for microRNA-155 detection by exonuclease III-aided cascade target recycling. J. Electroanal. Chem. 2020, 878, 114669. [Google Scholar] [CrossRef]
- Arduini, F.; Cinti, S.; Mazzaracchio, V.; Scognamiglio, V.; Amine, A.; Moscone, D. Carbon black as an outstanding and affordable nanomaterial for electrochemical (bio)sensor design. Biosens. Bioelectron. 2020, 156, 112033. [Google Scholar] [CrossRef]
- Hou, C.; Tang, W.; Zhang, C.; Wang, Y.; Zhu, N. A novel and sensitive electrochemical sensor for bisphenol A determination based on carbon black supporting ferroferric oxide nanoparticles. Electrochim. Acta 2014, 144, 324–331. [Google Scholar] [CrossRef]
- Kuzin, Y.; Kappo, D.; Porfireva, A.; Shurpik, D.; Stoikov, I.; Evtugyn, G.; Hianik, T. Electrochemical DNA Sensor Based on Carbon Black—Poly(Neutral Red) Composite for Detection of Oxidative DNA Damage. Sensors 2018, 18, 3489. [Google Scholar] [CrossRef] [Green Version]
- Rashid, J.I.A.; Yusof, N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: A review. Sens. Bio-Sens. Res. 2017, 16, 19–31. [Google Scholar] [CrossRef]
- Abedi, R.; Raoof, J.B.; Hashkavayi, A.B.; Asghari, M.; Azimi, R.; Hejazi, M.S. A novel genosensor based on Fe3O4@SiO2/DABCO-modified screen-printed graphite electrode for detection of prostate cancer gene sequence hybridization. J. Iran. Chem. Soc. 2022, 19, 2631–2640. [Google Scholar] [CrossRef]
- Shoaie, N.; Forouzandeh, M.; Omidfar, K. Voltammetric determination of the Escherichia coli DNA using a screen-printed carbon electrode modified with polyaniline and gold nanoparticles. Mikrochim. Acta 2018, 185, 217. [Google Scholar] [CrossRef] [PubMed]
- Yammouri, G.; Mohammadi, H.; Amine, A. A Highly Sensitive Electrochemical Biosensor Based on Carbon Black and Gold Nanoparticles Modified Pencil Graphite Electrode for microRNA-21 Detection. Chem. Afr. 2019, 2, 291–300. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zhang, B.; Lin, X.; Weng, W. Hybridization biosensor based on the covalent immobilization of probe DNA on chitosan–mutiwalled carbon nanotubes nanocomposite by using glutaraldehyde as an arm linker. Sens. Actuators B Chem. 2011, 156, 599–605. [Google Scholar] [CrossRef]
- Yola, M.L.; Eren, T.; Atar, N. A novel and sensitive electrochemical DNA biosensor based on Fe@Au nanoparticles decorated graphene oxide. Electrochim. Acta 2014, 125, 38–47. [Google Scholar] [CrossRef]
- Lin, X.-H.; Wu, P.; Chen, W.; Zhang, Y.-F.; Xia, X.-H. Electrochemical DNA biosensor for the detection of short DNA species of Chronic Myelogenous Leukemia by using methylene blue. Talanta 2007, 72, 468–471. [Google Scholar] [CrossRef]
- Nimse, S.B.; Song, K.; Sonawane, M.D.; Sayyed, D.R.; Kim, T. Immobilization techniques for microarray: Challenges and applications. Sensors 2014, 14, 22208–22229. [Google Scholar] [CrossRef] [Green Version]
- El Aamri, M.; Mohammadi, H.; Amine, A. Development of a Novel Electrochemical Sensor Based on Functionalized Carbon Black for the Detection of Guanine Released from DNA Hydrolysis. Electroanalysis 2022, 36, e6610. [Google Scholar] [CrossRef]
- Du, P.; Li, H.; Mei, Z.; Liu, S. Electrochemical DNA biosensor for the detection of DNA hybridization with the amplification of Au nanoparticles and CdS nanoparticles. Bioelectrochemistry 2009, 75, 37–43. [Google Scholar] [CrossRef]
- Mandli, J.; Mohammadi, H.; Amine, A. Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles. Bioelectrochemistry 2017, 116, 17–23. [Google Scholar] [CrossRef]
- Pothipor, C.; Aroonyadet, N.; Bamrungsap, S.; Jakmunee, J.; Ounnunkad, K. A highly sensitive electrochemical microRNA-21 biosensor based on intercalating methylene blue signal amplification and a highly dispersed gold nanoparticles/graphene/polypyrrole composite. Analyst 2021, 146, 2679–2688. Available online: https://pubs.rsc.org/en/content/articlehtml/2021/an/d1an00116g (accessed on 23 September 2022). [CrossRef]
- Aamri, M.E.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical biosensors for detection of MicroRNA as a cancer biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, J.; Han, X.; Cui, Y.; Wang, W. Electrochemical DNA biosensor fabrication with hollow gold nanospheres modified electrode and its enhancement in DNA immobilization and hybridization. Biosens. Bioelectron. 2010, 25, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yang, T.; Li, X.; Jiao, K. Fabrication of DNA/graphene/polyaniline nanocomplex for label-free voltammetric detection of DNA hybridization. Talanta 2012, 88, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Z.; Yang, M.; Li, J.; Zheng, F.; Shen, G.; Yu, R. Electrical detection of deoxyribonucleic acid hybridization based on carbon-nanotubes/nano zirconium dioxide/chitosan-modified electrodes. Anal. Chim. Acta 2007, 584, 268–274. [Google Scholar] [CrossRef]
- Xie, S.; Dong, Y.; Yuan, Y.; Chai, Y.; Yuan, R. Ultrasensitive Lipopolysaccharides Detection Based on Doxorubicin Conjugated N-(Aminobutyl)-N-(ethylisoluminol) as Electrochemiluminescence Indicator and Self-Assembled Tetrahedron DNA Dendrimers as Nanocarriers. Anal. Chem. 2016, 88, 5218–5224. [Google Scholar] [CrossRef]
- Yesil, M.; Donmez, S.; Arslan, F. Development of an electrochemical DNA biosensor for detection of specific Mycobacterium tuberculosis sequence based on poly(L-glutamic acid) modified electrode. J. Chem. Sci. 2016, 128, 1823–1829. [Google Scholar] [CrossRef]
- Kerman, K.; Ozkan, D.; Kara, P.; Meric, B.; Gooding, J.; Ozsoz, M. Voltammetric determination of DNA hybridization using methylene blue and self-assembled alkanethiol monolayer on gold electrodes. Anal. Chim. Acta 2002, 462, 39–47. [Google Scholar] [CrossRef]
- Erdem, A.; Kerman, K.; Meric, B.; Akarca, U.S.; Ozsoz, M. Novel hybridization indicator methylene blue for the electrochemical detection of short DNA sequences related to the hepatitis B virus. Anal. Chim. Acta 2000, 422, 139–149. [Google Scholar] [CrossRef]
- Nascimento, G.A.; Souza, E.V.; Campos-Ferreira, D.S.; Arruda, M.S.; Castelletti, C.H.; Wanderley, M.S.; Ekert, M.H.; Bruneska, D.; Lima-Filho, J.L. Electrochemical DNA biosensor for bovine papillomavirus detection using polymeric film on screen-printed electrode. Biosens. Bioelectron. 2012, 38, 61–66. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Gangopadhyay, R.; De, A. Highly sensitive electrochemical biosensor for glucose, DNA and protein using gold-polyaniline nanocomposites as a common matrix. Sens. Actuators B Chem. 2014, 190, 348–356. [Google Scholar] [CrossRef]
- Silvestrini, M.; Fruk, L.; Moretto, L.M.; Ugo, P. Detection of DNA Hybridization by Methylene Blue Electrochemistry at Activated Nanoelectrode Ensembles. J. Nanosci. Nanotechnol. 2015, 15, 3437–3442. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; Sklenar, H.; Lavery, A.R.; Röder, B. Methylene Blue Binding to DNA with Alternating GC Base Sequence: A Modeling Study. J. Am. Chem. Soc. 2000, 122, 2860–2866. [Google Scholar] [CrossRef]
- Liu, S.F.; Li, X.H.; Li, Y.C.; Li, Y.F.; Li, J.R.; Jiang, L. The influence of gold nanoparticle modified electrode on the structure of mercaptopropionic acid self-assembly monolayer. Electrochim. Acta 2005, 51, 427–431. [Google Scholar] [CrossRef]
- Saprigin, A.V.; Thomas, C.W.; Dulcey, C.S.; Patterson, C.H., Jr.; Spector, M.S. Spectroscopic quantification of covalently immobilized oligonucleotides. Surf. Interface Anal. 2005, 37, 24–32. [Google Scholar] [CrossRef]
- Xie, J.-K.; Jiao, K.; Liu, H.; Wang, Q.-X.; Liu, S.-F.; Fu, X. DNA Electrochemical Sensor Based on PbSe Nanoparticle for the Sensitive Detection of CaMV35S Gene Sequence. Chin. J. Anal. Chem. 2008, 36, 874–878. [Google Scholar] [CrossRef]
- Singh, R.; Sumana, G.; Verma, R.; Sood, S.; Sood, K.; Gupta, R.K.; Malhotra, B. Fabrication of Neisseria gonorrhoeae biosensor based on chitosan–MWCNT platform. Thin Solid Films 2010, 519, 1135–1140. [Google Scholar] [CrossRef]
- Erdem, A.; Papakonstantinou, P.; Murphy, H.; McMullan, M.; Karadeniz, H.; Sharma, S. Streptavidin Modified Carbon Nanotube Based Graphite Electrode for Label-Free Sequence Specific DNA Detection. Electroanalysis 2010, 22, 611–617. [Google Scholar] [CrossRef]
- Campos-Ferreira, D.S.; Nascimento, G.A.; Souza, E.V.; Souto-Maior, M.A.; Arruda, M.S.; Zanforlin, D.M.; Ekert, M.H.; Bruneska, D.; Lima-Filho, J.L. Electrochemical DNA biosensor for human papillomavirus 16 detection in real samples. Anal. Chim. Acta 2013, 804, 258–263. [Google Scholar] [CrossRef]
- Mohammadi, H.; Yammouri, G.; Amine, A. Current advances in electrochemical genosensors for detecting microRNA cancer markers. Curr. Opin. Electrochem. 2019, 16, 96–105. [Google Scholar] [CrossRef]
- Wang, Y.; Zulpya, M.; Zhang, X.; Xu, S.; Sun, J.; Dong, B. Recent Advances of Metal-Organic Frameworks-based Nanozymes for Bio-applications. Chem. Res. Chin. Univ. 2022, 38, 1–20. [Google Scholar] [CrossRef]
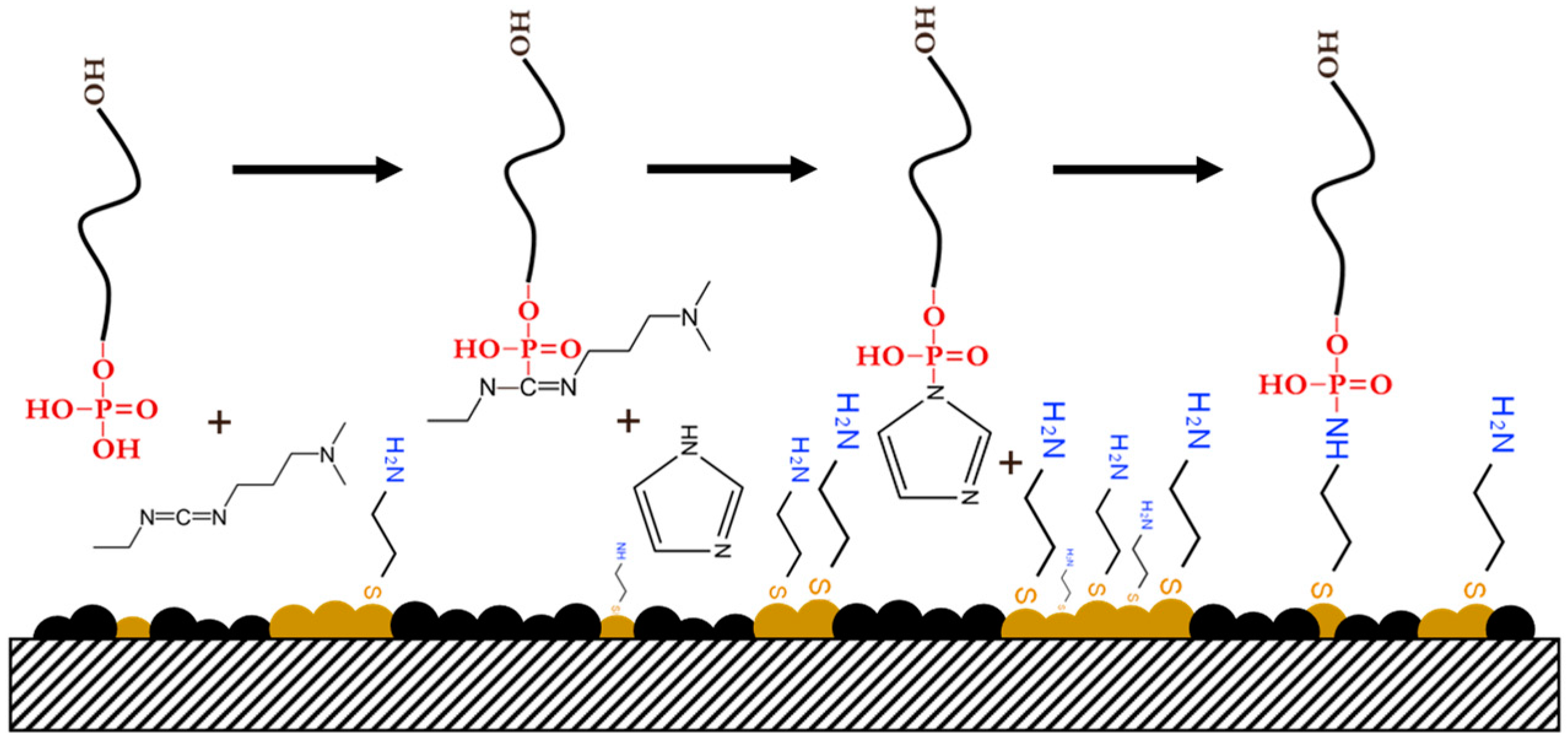

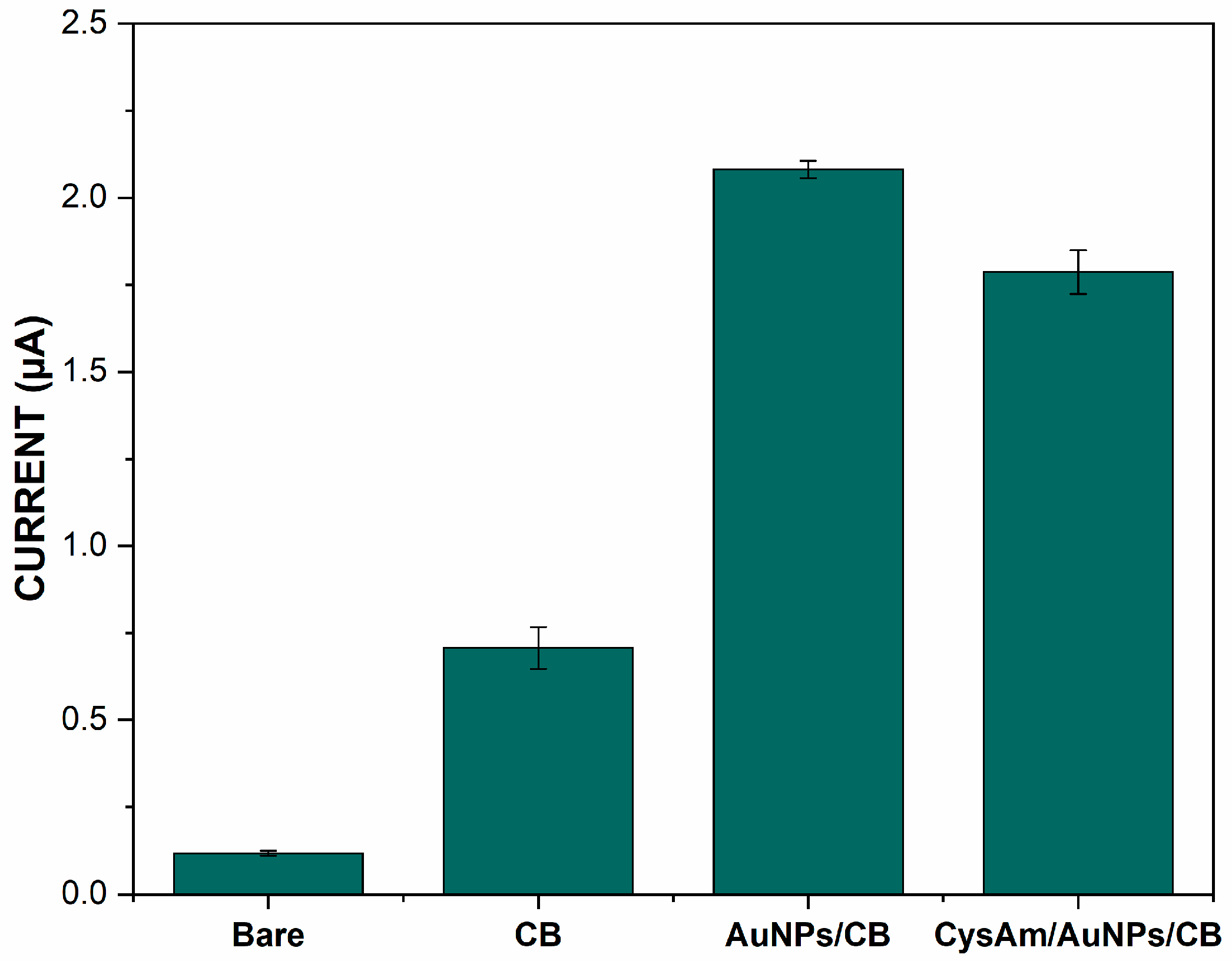
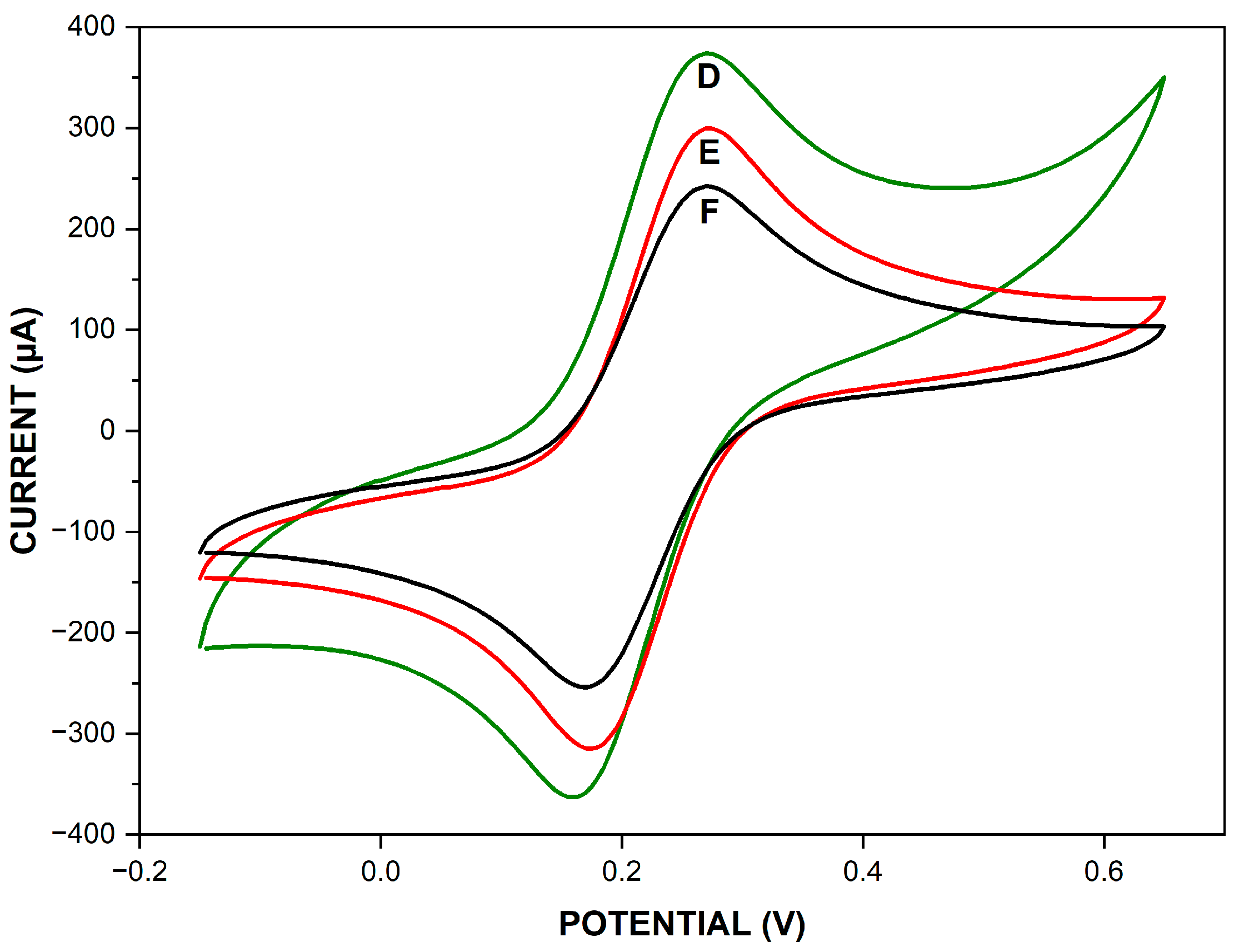
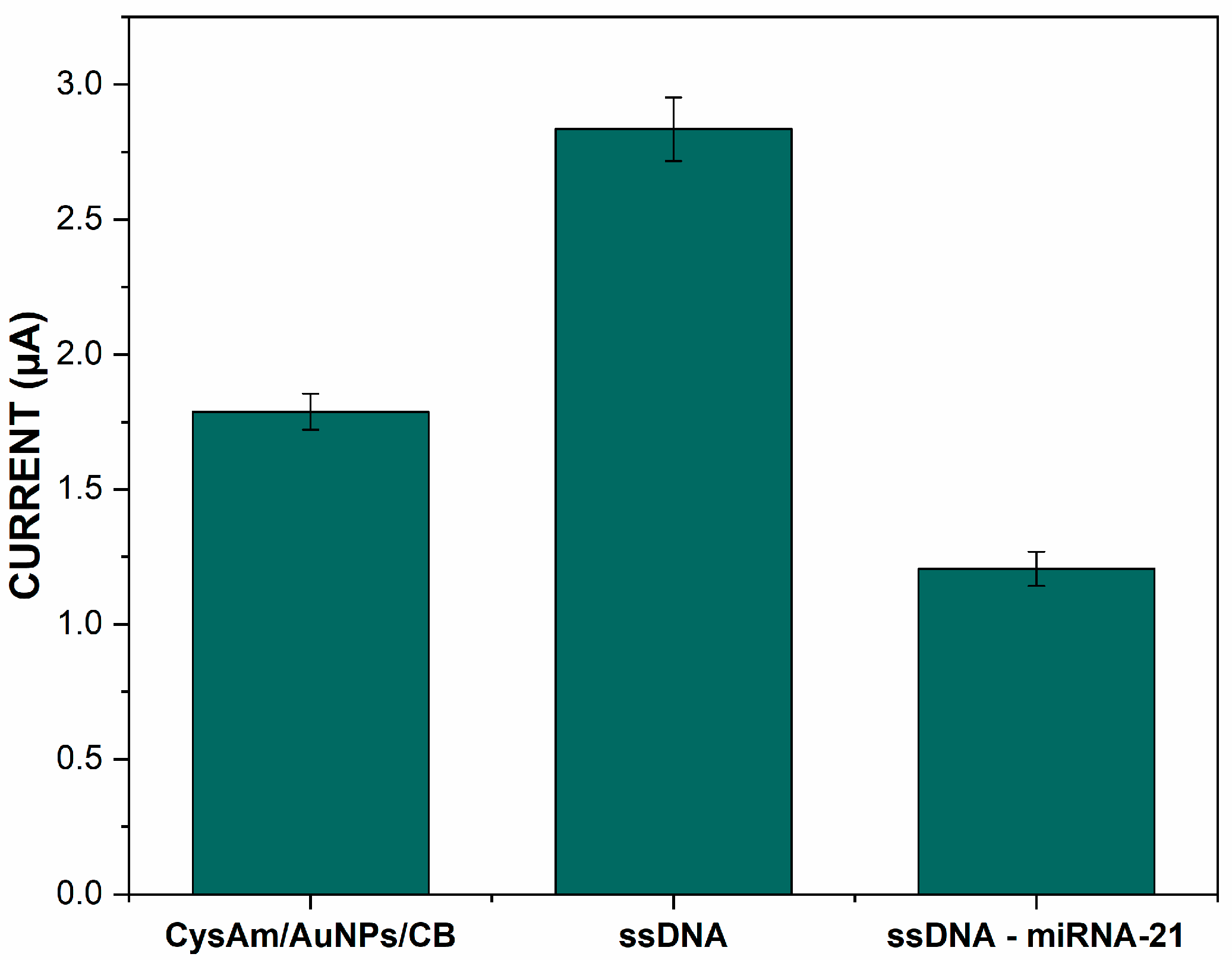

| Immobilization Matrix | Immobilization Technique | Chemical Interaction | DNA Modification | Advantages | Drawbacks | LOD (M) | Ref. |
|---|---|---|---|---|---|---|---|
| Poly-L-Lysine | Adsorption | Electrostatic adsorption | No modification | Simple, fast, and direct (no linker molecules) | Desorption risk and random orientation | 4.4 × 10−9 | [49] |
| Chitosan | 1.6 × 10−11 | [55] | |||||
| Avidin-Chitosan/MWCNTs | Avidin-Biotin | Avidin-Biotin affinity | Biotin-modified DNA | Improved orientation, high specificity, and reversible | Use of biocompatible linker and expensive | 1.0 × 10−16 | [56] |
| Avidin-MWCNTs | 1.5 × 10−7 | [57] | |||||
| GTD/Chitosan-MWCNTs | Covalent | Aldimine bond | Amine-modified DNA | Good orientation, high binding strength, and adaptative linker | Use of linker molecules and expensive | 8.5 × 10−14 | [33] |
| Cysteine film | Au-S bond | Thiol-modified DNA | 18 × 10−8 | [58] | |||
| CysAm/AuNPs/CB | Covalent | Phosphoramidate bond | No modification | Good orientation, high binding strength, and no linker molecules needed | Reaction time and irreversible | 1 × 10−9 | This work |
| Added (nM) | Detected (nM) | ΔI (µA) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 10 | 9.84 | 0.443 | 98.4 | 2.15 |
| 50 | 47.37 | 0.942 | 94.74 | 2.75 |
| 125 | 127.66 | 1.309 | 102.13 | 2.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moustakim, H.; Mohammadi, H.; Amine, A. Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer. Sensors 2022, 22, 9420. https://doi.org/10.3390/s22239420
Moustakim H, Mohammadi H, Amine A. Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer. Sensors. 2022; 22(23):9420. https://doi.org/10.3390/s22239420
Chicago/Turabian StyleMoustakim, Hamza, Hasna Mohammadi, and Aziz Amine. 2022. "Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer" Sensors 22, no. 23: 9420. https://doi.org/10.3390/s22239420





