Segmentation Performance Comparison Considering Regional Characteristics in Chest X-ray Using Deep Learning
Abstract
:1. Introduction
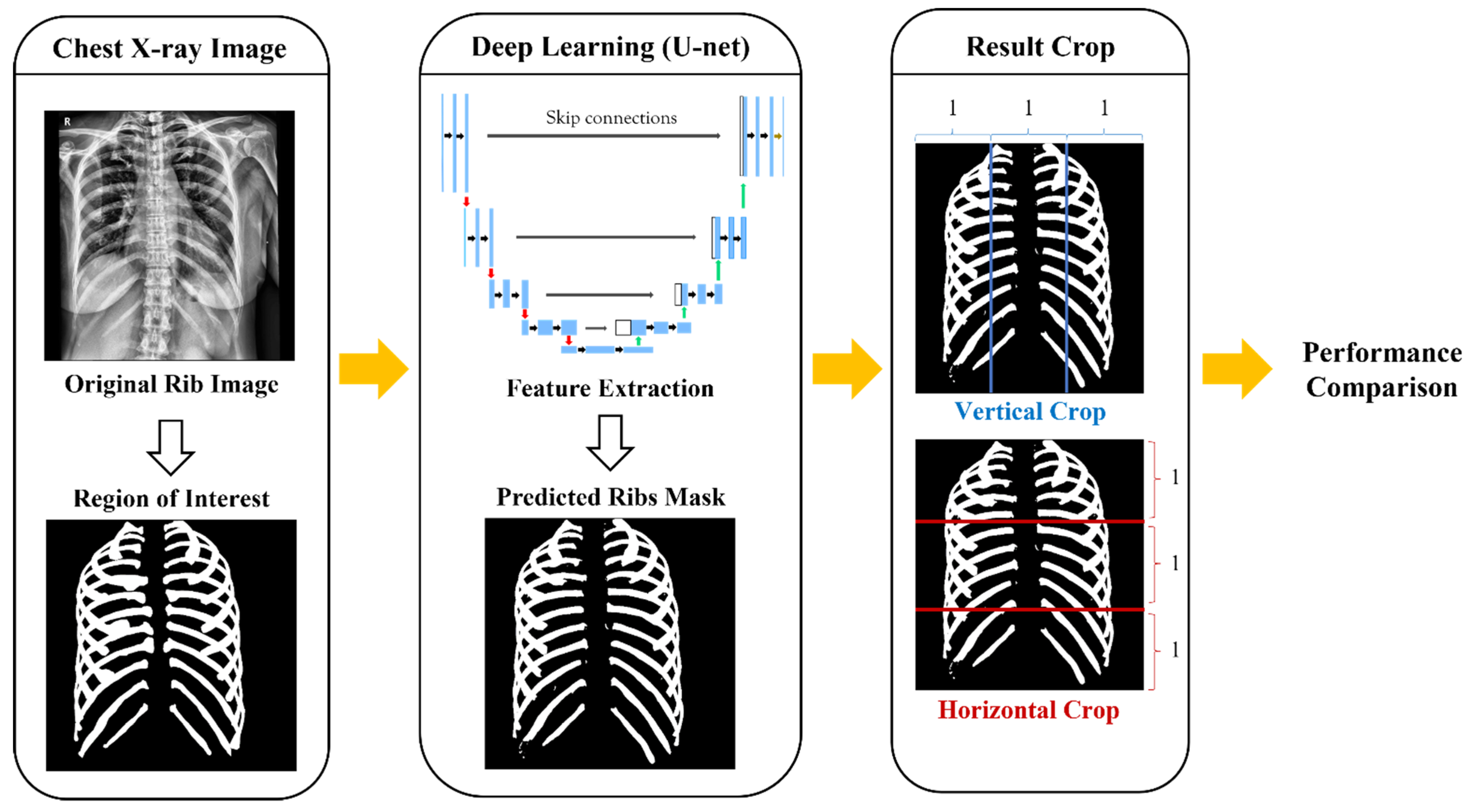
2. Materials and Methods
2.1. Data Collection and Annotation
2.2. Data Pre-Processing
2.3. U-Net Architecture
2.4. Criteria of Dividing Ribs
3. Results
3.1. Whole Ribs Segmentation Model Performance
3.2. Comparison Performance Difference between Medial and Lateral Ribs
3.3. Comparison Performance Difference between Superior, Middle, and Inferior Ribs
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pelicano, A.C.; Gonçalves, M.C.T.; Godinho, D.M.; Castela, T.; Orvalho, M.L.; Ara, N.A.M.; Porter, E.; Conceiç, R.C. Development of 3D MRI-Based Anatomically Realistic Models of Breast Tissues and Tumors for Microwave Imaging Diagnosis. Sensors 2021, 21, 8265. [Google Scholar] [CrossRef]
- Tsoumakidou, M.; Chrysofakis, G.; Tsiligianni, I.; Maltezakis, G.; Siafakas, N.M.; Tzanakis, N. A Prospective Analysis of 184 Hemoptysis Cases—Diagnostic Impact of Chest X-ray, Computed Tomography, Bronchoscopy. Respiration 2006, 73, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Carmody, D.P.; Nodine, C.F.; Kundel, H.L. An Analysis of Perceptual and Cognitive Factors in Radiographic Interpretation. Perception 1980, 9, 339–344. [Google Scholar] [CrossRef]
- O’Connor, S.D.; Yao, J.; Summers, S.R. Lytic Metastases in Thoracolumbar Spine: Computer-Aided Detection at CT—Preliminary Study. Radiology 2007, 242, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.H.; Romney, B.M.; Missed, L.S. Bronchogenic Carcinoma: Radiographic Findings in 27 Patients with a Potentially Resectable Lesion Evident in Retrospect. Radiology 1992, 182, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Doi, K.; Kobayashi, T.; MacMahon, H.; Giger, M.L. Development of an Improved CAD Scheme for Automated Detection of Lung Nodules in Digital Chest Images. Med. Phys. 1997, 24, 1395–1403. [Google Scholar] [CrossRef]
- Shiraishi, J.; Li, Q.; Suzuki, K.; Engelmann, R.; Doi, K. Computer-Aided Diagnostic Scheme for the Detection of Lung Nodules on Chest Radiographs: Localized Search Method Based on Anatomical Classification. Med. Phys. 2006, 33, 2642–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.; Kumar, N.; Patil, A. MoNuSAC2020: A Multi-Organ Nuclei Segmentation and Classification Challenge. IEEE Trans. Med. Imaging 2021, 40, 3413–3423. [Google Scholar] [CrossRef]
- Greenwald, N.F.; Miller, G.; Moen, E.; Kong, A.; Kagel, A.; Dougherty, T.; Fullaway, C.S.; McIntosh, B.J.; Leow, K.X.; Schwartz, M.S. Whole-Cell Segmentation of Tissue Images with Human-Level Performance Using Large-Scale Data Annotation and Deep Learning. Nat. Biotechnol. 2021, 40, 1–29. [Google Scholar] [CrossRef]
- Staal, J.; van Ginneken, B.; Viergever, M.A. Automatic Rib Segmentation and Labeling in Computed Tomography Scans Using a General Framework for Detection, Recognition and Segmentation of Objects in Volumetric Data. Med. Image Anal. 2007, 11, 35–46. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Song, H.-J.; Lee, E.-B.; Jo, H.-J.; Park, S.-Y.; Kim, S.-Y.; Kim, H.-J.; Hong, J.-W. Evaluation of Classification and Accuracy in Chest X-ray Images Using Deep Learning with Convolution Neural Network. J. Korean Soc. Radiol. 2020, 14, 39–44. [Google Scholar]
- Zhang, Y.; Gorriz, J.M.; Dong, Z. Deep Learning in Medical Image Analysis. J. Imaging 2021, 7, 74. [Google Scholar] [CrossRef]
- Sharma, H.B.; Panigrahi, S.; Sarmah, A.K.; Dubey, B.K. ULNet for the Detection of Coronavirus (COVID-19) from Chest X-ray Images. Sci. Total Environ. 2021, 137, 104834. [Google Scholar]
- Al Arif, S.M.M.R.; Knapp, K.; Slabaugh, G. Fully Automatic Cervical Vertebrae Segmentation Framework for X-ray Images. Comput. Methods Programs Biomed. 2018, 157, 95–111. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; Mota, V.; Machado, A.M.; dos Santos, J.A. From 3D to 2D: Transferring Knowledge for Rib Segmentation in Chest X-rays. Pattern Recognit. Lett. 2020, 140, 10–17. [Google Scholar] [CrossRef]
- Kundel, H.L.; Nodine, N.C. Interpreting Chest Radiographs without Visual Search. Radiology 1975, 116, 527–532. [Google Scholar] [CrossRef]
- Liang, C.; Xin, S. Research Status and Prospects of Deep Learning in Medical Images. In Proceedings of the 2020 International Conference on Communications, Information System and Computer Engineering (CISCE), Kuala Lumpur, Malaysia, 3–5 July 2020. [Google Scholar]
- Wang, W.; Feng, H.; Bu, Q.; Cui, L.; Xie, Y.; Zhang, A.; Feng, J.; Zhu, Z.; Chen, Z. MDU-Net: A Convolutional Network for Clavicle and Rib Segmentation from a Chest Radiograph. J. Healthc. Eng. 2020, 2020, 9. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Yadav, S.; Shukla, S. Analysis of K-Fold Cross-Validation over Hold-Out Validation on Colossal Datasets for Quality Classification. In Proceedings of the 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016. [Google Scholar]
- Kim, Y.J.; Ganbold, B.; Kim, K.G. Web-Based Spine Segmentation Using Deep Learning in Computed Tomography Images. Healthc. Inform. Res. 2020, 26, 61–67. [Google Scholar] [CrossRef]
- Horng, M.H.; Kuok, C.P.; Fu, M.J.; Lin, C.J.; Sun, Y.N. Cobb Angle Measurement of Spine from X-ray Images Using Convolutional Neural Network. Comput. Math. Methods Med. 2019, 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [Green Version]
- Albishri, A.A.; Shah, S.J.H.; Lee, Y. CU-Net: Cascaded U-Net Model for Automated Liver and Lesion Segmentation and Summarization. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. Importance of skip connections in biomedical image segmentation. arXiv 2016, arXiv:1608.04117. [Google Scholar] [CrossRef] [Green Version]
- Seibert, J.A. X-ray Imaging Physics for Nuclear Medicine Technologists. J. Nucl. Med. Technol. 2004, 32, 139–147. [Google Scholar]
- Wolde Hawariat, B.Y.; Fenta, B.D.; Gebreselassie, H.A. Subcutaneous Chest Wall Hamartoma: Case Report. J. Pediatr. Surg. Case Rep. 2021, 73, 102009. [Google Scholar] [CrossRef]
- Nirula, R.; Diaz, J.J.; Trunkey, D.D.; Mayberry, J.C. Rib Fracture Repair: Indications, Technical Issues, and Future Directions. World J. Surg. 2009, 33, 14–22. [Google Scholar] [CrossRef]
- Zhang, R.; Du, L.; Xiao, Q.; Liu, J. Comparison of Backbones for Semantic Segmentation Network. J. Phys. Conf. Ser. 2020, 1544, 012196. [Google Scholar] [CrossRef]
- Cai, W.; Wei, Z.; Song, Y.; Li, M.; Yang, X. Residual-Capsule Networks with Threshold Convolution for Segmentation of Wheat Plantation Rows in UAV Images. Multimed. Tools Appl. 2021, 80, 32131–32147. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Alquraan, H.; Qasmieh, I.A. Segmented and Non-Segmented Skin Lesions Classification Using Transfer Learning and Adaptive Moment Learning Rate Technique Using Pretrained Convolutional Neural Network. J. Biomim. Biomater. Biomed. Eng. 2019, 42, 67–78. [Google Scholar]
- Chen, J.; Wan, Z.; Zhang, J.; Li, W.; Chen, Y.; Li, Y.; Duan, Y. Medical Image Segmentation and Reconstruction of Prostate Tumor Based on 3D AlexNet. Comput. Methods Programs Biomed. 2021, 200, 105878. [Google Scholar] [CrossRef] [PubMed]






| Unit Level | Conv Layer | Filter | Activation Function | Output Size | |
|---|---|---|---|---|---|
| Input | 1024 × 1024 × 1 | ||||
| Encoding | Level 1 | Conv 1 | 3 × 3/32 | ReLu | 1024 × 1024 ×32 |
| Conv 2 | 3 × 3/32 | 1024 × 1024 × 32 | |||
| Level 2 | Conv 3 | 3 × 3/64 | ReLu | 512 × 512 × 64 | |
| Conv 4 | 3 × 3/64 | 512 × 512 × 64 | |||
| Level 3 | Conv 5 | 3 × 3/128 | ReLu | 256 × 256 × 128 | |
| Conv 6 | 3 × 3/128 | 256 × 256 × 128 | |||
| Level 4 | Conv 7 | 3 × 3/256 | ReLu | 128 × 128 × 256 | |
| Conv 8 | 3 × 3/256 | 128 × 128 × 256 | |||
| Bridge | Level 5 | Conv 9 | 3 × 3/512 | ReLu | 64 × 64 × 512 |
| Conv 10 | 3 × 3/512 | 64 × 64 × 512 | |||
| Decoding | Level 6 | Conv 11 | 3 × 3/256 | ReLu | 128 × 128 × 256 |
| Conv 12 | 3 × 3/256 | 128 × 128 × 256 | |||
| Level 7 | Conv 13 | 3 × 3/128 | ReLu | 256 × 256 × 128 | |
| Conv 14 | 3 × 3/128 | 256 × 256 × 128 | |||
| Level 8 | Conv 15 | 3 × 3/64 | ReLu | 512 × 512 × 64 | |
| Conv 16 | 3 × 3/64 | 512 × 512 × 64 | |||
| Level 9 | Conv 17 | 3 × 3/32 | ReLu | 1024 × 1024 × 32 | |
| Conv 18 | 3 × 3/32 | 1024 × 1024 × 32 | |||
| Output | Conv 19 | 1 × 1 | Sigmoid | 1024 × 1024 × 1 |
| Folds | Precision (%) | Recall (%) | Sensitivity (%) | DSC (%) |
|---|---|---|---|---|
| Fold-1 | 92.06 | 87.54 | 87.17 | 88.95 |
| Fold-2 | 92.72 | 87.61 | 87.62 | 89.1 |
| Fold-3 | 97.74 | 88.44 | 87.41 | 89.74 |
| Fold-4 | 97.76 | 90.14 | 89.13 | 91.08 |
| Fold-5 | 87.25 | 89.04 | 89.39 | 89.39 |
| Average | 93.51 ± 0.04 | 88.56 ± 0.86 | 88.14 ± 0.05 | 89.65 ± 0.04 |
| Precision (%) | Recall (%) | Sensitivity (%) | DSC (%) | |
|---|---|---|---|---|
| Medial Ribs | 89.76 ± 0.07 | 82.66 ± 0.82 | 82.53 ± 0.08 | 84.76 ± 0.04 |
| Lateral Ribs | 95.22 ± 0.03 | 90.89 ± 0.9 | 91.03 ± 0.04 | 92.03 ± 0.03 |
| Precision (%) | Recall (%) | Sensitivity (%) | DSC (%) | |
|---|---|---|---|---|
| Superior Ribs | 92.69 ± 0.04 | 88.92 ± 0.88 | 88.92 ± 0.05 | 89.94 ± 0.04 |
| Middle Ribs | 94.11 ± 0.04 | 90.31 ± 0.90 | 89.88 ± 0.05 | 91.01 ± 0.04 |
| Inferior Ribs | 93.45 ± 0.04 | 83.53 ± 0.83 | 83.294 ± 0.1 | 85.8 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.M.; Kim, Y.J.; Kim, K.G. Segmentation Performance Comparison Considering Regional Characteristics in Chest X-ray Using Deep Learning. Sensors 2022, 22, 3143. https://doi.org/10.3390/s22093143
Lee HM, Kim YJ, Kim KG. Segmentation Performance Comparison Considering Regional Characteristics in Chest X-ray Using Deep Learning. Sensors. 2022; 22(9):3143. https://doi.org/10.3390/s22093143
Chicago/Turabian StyleLee, Hyo Min, Young Jae Kim, and Kwang Gi Kim. 2022. "Segmentation Performance Comparison Considering Regional Characteristics in Chest X-ray Using Deep Learning" Sensors 22, no. 9: 3143. https://doi.org/10.3390/s22093143






