Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance
Abstract
1. Introduction
1.1. Related Work
1.2. Contributions and Structure
- Introduction of a new application for specific deep CNNs to classify the same-limb MI, with the aim of improving classification accuracy beyond the state-of-the art;
- Benchmarking and in-depth performance analysis for various classification methods, including the commonly used and the more architecturally complex newly applied methods;
- In-depth statistical analyses of the effects of different guidance techniques (visual guidance or a combination of visual and vibrotactile guidance) and different data preprocessing on classification accuracy for all observed methods.
2. Materials and Methods
2.1. Datasets
2.1.1. ULM Dataset
2.1.2. KGU Dataset
2.2. Data Preprocessing
- Bad trials, based on amplitude threshold and artifact presence, were rejected using the EEGLAB Matlab toolbox [54].
- Independent component analysis (ICA) [54,55] was performed separately for each participant. For the ULM dataset, it was performed for 31 EEG channels (yielding 31 independent components). The remaining 3 EOG channels were used for artifact removal. For the KGU dataset, it was performed for 61 EEG channels (yielding 61 independent components). In this case, the EOG channels were also used for artifact removal. For both datasets, only relevant independent components (IC) were retained, using SASICA [56] and manual IC rejection.
- The data were further filtered (fourth-order zero-phase Butterworth filter) in the bands of interest, specifically 0.2–5 Hz for low-frequency features and 1–40 Hz for broad-frequency features. Only relevant MI periods were epoched for classification (from s to s for the ULM dataset, as shown in Figure 2, and from s to s for the KGU dataset, as shown in Figure 3).
- The features were then further downsampled to 20 Hz for low-frequency features and to 100 Hz for broad-frequency features.
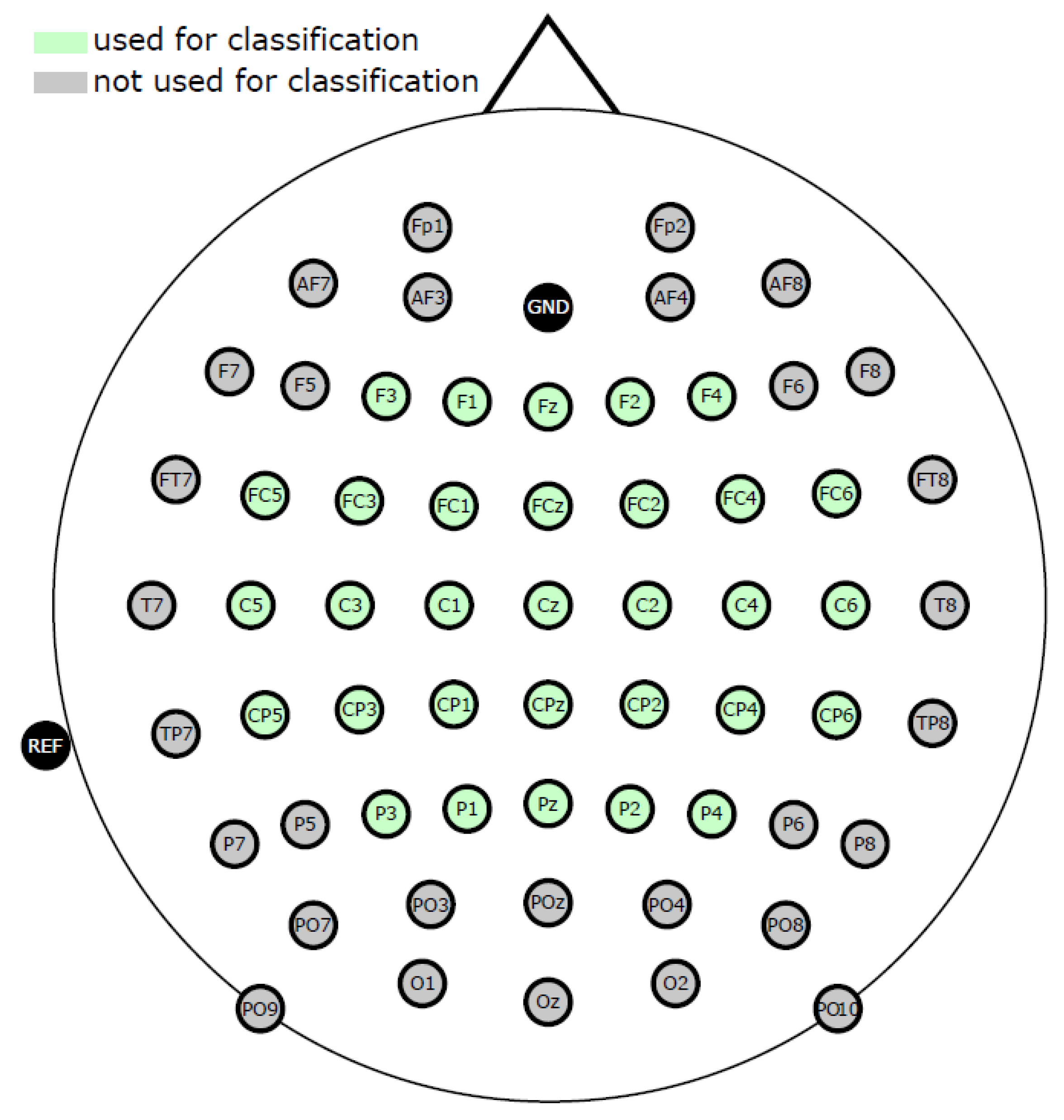
- In total, 31 relevant channels around the motor-related area were selected for further preprocessing and analysis (Figure 1).
2.3. Classification
2.3.1. Shrinkage Linear Discriminant Analysis
- Calculate the shrinkage LDA coefficients, i.e., the weights that define the linear boundary between the different classes.
- Calculate the class means for each group.
- Calculate the pooled within-class covariance matrix, which is a weighted sum of the sample covariance matrices for each group.
- Calculate the shrinkage covariance matrix using the formula:where S is the sample covariance matrix, D is the diagonal matrix, and is the shrinkage parameter.
- Calculate the inverse of the pooled within-class covariance matrix.
- Calculate the discriminant value for each observation using the formula:where x is the feature vector, is the inverse of the pooled within-class covariance matrix, and and are the means of the two classes.
- Classify each observation based on the sign of the discriminant value. If , the observation is classified as belonging to the first class. If , then the observation is classified as belonging to the second class.
2.3.2. Support Vector Machine
- Given a set of training data, the kernel SVM selects a subset of data points as support vectors, i.e., the points closest to the decision boundary in higher-dimensional space.
- The kernel SVM then finds the hyperplane that maximizes the distance between the support vectors of each class in the higher-dimensional space.
- To classify new observations, the kernel SVM maps them into higher-dimensional space using the kernel function, projects them onto the hyperplane, and assigns them to the class on the corresponding side. The sign of the projection determines the class of the observation.
2.3.3. Random Forest
- Randomly sample the training data with replacement (bootstrap) to create multiple datasets (or decision trees) of the same size as the original dataset.
- For each dataset, randomly select a subset of the input features to use for building the DT.
- Build a DT for each dataset using the selected features and a splitting criterion.
- Repeat steps 1–3 to create a forest of DT.
- To make a prediction for a new sample, pass it through all the DTs in the forest and average their predictions (for regression tasks) or take the majority vote (for classification tasks).
2.3.4. VGG-19
2.3.5. ResNet-101
2.3.6. DenseNet-169
3. Results and Discussion
3.1. Comparison of Classification Methods and Preprocessing Frequency Bands with the ULM Dataset
- Mean classification accuracy differed statistically significantly between observed methods: . Post-hoc analysis with a Bonferroni adjustment confirmed that ResNet-101 statistically achieves the best results among all competitors (). The complete results of post hoc pairwise comparisons are given in Table 2.
- There is no significant effect of on classification accuracy: . In other words, the difference in classification accuracy when preprocessing the ULM dataset using a low-frequency band (0.2–5 Hz) and using a broad-frequency band (1–40 Hz) is not statistically significant.
- The interaction between the factors is not statistically significant: .
3.2. Comparison of Classification Methods, Guidance Types, and Preprocessing Frequency Bands with the KGU Dataset
- A significant effect of the on classification accuracy was again found: . Similar to the case of the ULM dataset, ResNet-101 achieved the best classification accuracy (mean value of ). By far the worst accuracy, however, was obtained with the RF method (mean value of ). The results of the post hoc pairwise comparisons with Bonferroni adjustment are shown in Table 4.
- A significant effect of on classification accuracy was also found: . Hence, the classification accuracy when using vibrotactile guidance (VtG, ) is higher than in the case where there is no such type of assistance (noVtG, ). Although this difference may seem negligible in absolute terms, it is still statistically significant.
- The third observed factor, , has a significant effect on classification accuracy as well: . If the data are preprocessed using a filter with a low-frequency band, a significantly higher accuracy is achieved () than with a broad-frequency band ().
- None of the interactions between the observed factors are statistically significant:
- –
- : .
- –
- : .
- –
- : .
- –
- : .

4. Conclusions
Limitations and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCI | Brain–computer interface |
| CNN | Convolutional neural network |
| DNN | Deep neural network |
| DT | Decision tree |
| ECoG | Electrocorticography |
| EE | Elbow extension |
| EEG | Electroencephalography |
| EF | Elbow flexion |
| EMG | Electromiogram |
| ERD | Event-related desynchronization |
| ERS | Event-related synchronization |
| fMRI | Functional magnetic resonance imaging |
| GCN | Graph convolutional neural network |
| IC | Independent component |
| ICA | Independent component analysis |
| KGU | Kinesthetic guidance (dataset) |
| LDA | Linear discriminant analysis |
| LFP | Local field potential |
| MEG | Magnetoencephalography |
| ME | Movement execution |
| MI | Motor imagery |
| NF | Neurofeedback |
| noVtG | MI condition without vibrotactile stimulation |
| QP | Quadratic programming |
| ResNet | Residual Network |
| RF | Random forest |
| RM | Repeated measures |
| sLDA | LDA with shrinkage regularization |
| SMR | Sensorimotor rhythms |
| SSVEP | Steady-state visual evoked potentials |
| SVM | Support vector machine |
| ULM | Upper limb movement (dataset) |
| VtG | MI condition with vibrotactile stimulation |
References
- He, B.; Baxter, B.; Edelman, B.J.; Cline, C.C.; Ye, W.W. Noninvasive Brain-Computer Interfaces Based on Sensorimotor Rhythms. Proc. IEEE 2015, 103, 907–925. [Google Scholar] [CrossRef] [PubMed]
- Farwell, L.; Donchin, E. Talking off the top of your head: Toward a mental prosthesis utilizing event-related brain potentials. Electroencephalogr. Clin. Neurophysiol. 1988, 70, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Kindermans, P.J.; Verschore, H.; Schrauwen, B. A Unified Probabilistic Approach to Improve Spelling in an Event-Related Potential-Based Brain–Computer Interface. IEEE Trans. Biomed. Eng. 2013, 60, 2696–2705. [Google Scholar] [CrossRef]
- Gu, Z.; Yu, Z.; Shen, Z.; Li, Y. An Online Semi-supervised Brain–Computer Interface. IEEE Trans. Biomed. Eng. 2013, 60, 2614–2623. [Google Scholar] [CrossRef]
- Postelnicu, C.C.; Talaba, D. P300-Based Brain-Neuronal Computer Interaction for Spelling Applications. IEEE Trans. Biomed. Eng. 2013, 60, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Middendorf, M.; McMillan, G.; Calhoun, G.; Jones, K. Brain-computer interfaces based on the steady-state visual-evoked response. IEEE Trans. Rehabil. Eng. 2000, 8, 211–214. [Google Scholar] [CrossRef]
- Kimura, Y.; Tanaka, T.; Higashi, H.; Morikawa, N. SSVEP-Based Brain–Computer Interfaces Using FSK-Modulated Visual Stimuli. IEEE Trans. Biomed. Eng. 2013, 60, 2831–2838. [Google Scholar] [CrossRef]
- Li, Y.; Pan, J.; Wang, F.; Yu, Z. A Hybrid BCI System Combining P300 and SSVEP and Its Application to Wheelchair Control. IEEE Trans. Biomed. Eng. 2013, 60, 3156–3166. [Google Scholar] [CrossRef]
- Yin, E.; Zhou, Z.; Jiang, J.; Chen, F.; Liu, Y.; Hu, D. A Speedy Hybrid BCI Spelling Approach Combining P300 and SSVEP. IEEE Trans. Biomed. Eng. 2014, 61, 473–483. [Google Scholar] [CrossRef]
- Wolpaw, J.R.; McFarland, D.J.; Neat, G.W.; Forneris, C.A. An EEG-based brain–computer interface for cursor control. Electroencephalogr. Clin. Neurophysiol. 1991, 78, 252–259. [Google Scholar] [CrossRef]
- Obermaier, B.; Muller, G.; Pfurtscheller, G. “Virtual keyboard” controlled by spontaneous EEG activity. IEEE Trans. Neural Syst. Rehabil. Eng. 2003, 11, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Perdikis, S.; Leeb, R.; Williamson, J.; Ramsay, A.; Tavella, M.; Desideri, L.; Hoogerwerf, E.; Al-Khodairy, A.; Murray-Smith, R.; Millán, J. Clinical evaluation of BrainTree, a motor imagery hybrid BCI speller. J. Neural Eng. 2014, 11, 036003. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, K.; Cassady, K.; Doud, A.; Shades, K.; Rogin, E.; He, B. Quadcopter control in three-dimensional space using a noninvasive motor imagery-based brain–computer interface. J. Neural Eng. 2013, 10, 046003. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Matsunaga, K.; Wang, H. Electroencephalogram-based control of an electric wheelchair. IEEE Trans. Robot. 2005, 21, 762–766. [Google Scholar] [CrossRef]
- Leeb, R.; Friedman, D.; Müller-Putz, G.; Scherer, R.; Slater, M.; Pfurtscheller, G. Self-paced (asynchronous) BCI control of a wheelchair in virtual environments: A case study with a tetraplegic. Comput. Intell. Neurosci. 2007, 2007, 079642. [Google Scholar] [CrossRef]
- Galán, F.; Nuttin, M.; Lew, E.; Ferrez, P.; Vanacker, G.; Philips, J.; del R. Millán, J. A brain-actuated wheelchair: Asynchronous and non-invasive Brain–computer interfaces for continuous control of robots. Clin. Neurophysiol. 2008, 119, 2159–2169. [Google Scholar] [CrossRef]
- Carlson, T.; del Millan, J.R. Brain-Controlled Wheelchairs: A Robotic Architecture. IEEE Robot. Autom. Mag. 2013, 20, 65–73. [Google Scholar] [CrossRef]
- Carlson, T.; Tonin, L.; Perdikis, S.; Leeb, R.; Millán, J.d.R. A hybrid BCI for enhanced control of a telepresence robot. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 3097–3100. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, S.; Bekyo, A.; Olsoe, J.; Baxter, B.; He, B. Noninvasive Electroencephalogram Based Control of a Robotic Arm for Reach and Grasp Tasks. Sci. Rep. 2016, 6, 38565. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Lopes da Silva, F. Event-related EEG/MEG synchronization and desynchronization: Basic principles. Clin. Neurophysiol. 1999, 110, 1842–1857. [Google Scholar] [CrossRef]
- Yuan, H.; He, B. Brain–Computer Interfaces Using Sensorimotor Rhythms: Current State and Future Perspectives. IEEE Trans. Bio-Med Eng. 2014, 61, 1425–1435. [Google Scholar] [CrossRef]
- Kobler, R.J.; Kolesnichenko, E.; Sburlea, A.I.; Müller-Putz, G.R. Distinct cortical networks for hand movement initiation and directional processing: An EEG study. NeuroImage 2020, 220, 117076. [Google Scholar] [CrossRef]
- Wolpaw, J.R.; McFarland, D.J. Multichannel EEG-based brain–computer communication. Electroencephalogr. Clin. Neurophysiol. 1994, 90, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Royer, A.S.; Doud, A.J.; Rose, M.L.; He, B. EEG Control of a Virtual Helicopter in 3-Dimensional Space Using Intelligent Control Strategies. IEEE Trans. Neural Syst. Rehabil. Eng. 2010, 18, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Doud, A.J.; Lucas, J.P.; Pisansky, M.T.; He, B. Continuous three-dimensional control of a virtual helicopter using a motor imagery based brain–computer interface. PLoS ONE 2011, 6, e26322. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, T.; Szarkowski, R.; Rios, C.; Ashe, J.; He, B. Negative covariation between task-related responses in alpha/beta-band activity and BOLD in human sensorimotor cortex: An EEG and fMRI study of motor imagery and movements. NeuroImage 2010, 49, 2596–2606. [Google Scholar] [CrossRef]
- Raza, H.; Rathee, D.; Zhou, S.M.; Cecotti, H.; Prasad, G. Covariate shift estimation based adaptive ensemble learning for handling non-stationarity in motor imagery related EEG-based brain–computer interface. Neurocomputing 2019, 343, 154–166. [Google Scholar] [CrossRef]
- Padfield, N.; Zabalza, J.; Zhao, H.; Masero, V.; Ren, J. EEG-based brain–computer interfaces using motor-imagery: Techniques and challenges. Sensors 2019, 19, 1423. [Google Scholar] [CrossRef]
- Miladinović, A.; Ajčević, M.; Jarmolowska, J.; Marusic, U.; Colussi, M.; Silveri, G.; Battaglini, P.P.; Accardo, A. Effect of power feature covariance shift on BCI spatial-filtering techniques: A comparative study. Comput. Methods Programs Biomed. 2021, 198, 105808. [Google Scholar] [CrossRef]
- Jeunet, C.; Glize, B.; McGonigal, A.; Batail, J.M.; Micoulaud-Franchi, J.A. Using EEG-based brain computer interface and neurofeedback targeting sensorimotor rhythms to improve motor skills: Theoretical background, applications and prospects. Neurophysiol. Clin. 2019, 49, 125–136. [Google Scholar] [CrossRef]
- Blankertz, B.; Sannelli, C.; Halder, S.; Hammer, E.M.; Kübler, A.; Müller, K.R.; Curio, G.; Dickhaus, T. Neurophysiological predictor of SMR-based BCI performance. NeuroImage 2010, 51, 1303–1309. [Google Scholar] [CrossRef]
- Kübler, A.; Nijboer, F.; Mellinger, J.; Vaughan, T.M.; Pawelzik, H.; Schalk, G.; McFarland, D.J.; Birbaumer, N.; Wolpaw, J.R. Patients with ALS can use sensorimotor rhythms to operate a brain–computer interface. Neurology 2005, 64, 1775–1777. [Google Scholar] [CrossRef] [PubMed]
- Dornhege, G.; del Millán, J.R.; Hinterberger, T.; McFarland, D.J.; Müller, K.R. (Eds.) Toward Brain–Computer Interfacing; MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Sainburg, R.L.; Ghilardi, M.F.; Poizner, H.; Ghez, C. Control of limb dynamics in normal subjects and patients without proprioception. J. Neurophysiol. 1995, 73, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Murguialday, A.; Schürholz, M.; Caggiano, V.; Wildgruber, M.; Caria, A.; Hammer, E.M.; Halder, S.; Birbaumer, N. Proprioceptive feedback and brain computer interface (BCI) based neuroprostheses. PLoS ONE 2012, 7, e47048. [Google Scholar] [CrossRef] [PubMed]
- Corbet, T.; Iturrate, I.; Pereira, M.; Perdikis, S.; del Millán, J.R. Sensory threshold neuromuscular electrical stimulation fosters motor imagery performance. NeuroImage 2018, 176, 268–276. [Google Scholar] [CrossRef]
- Hehenberger, L.; Sburlea, A.I.; Müller-Putz, G.R. Assessing the impact of vibrotactile kinaesthetic feedback on electroencephalographic signals in a center-out task. J. Neural Eng. 2020, 17, 44944–44950. [Google Scholar] [CrossRef]
- Hehenberger, L.; Batistic, L.; Sburlea, A.I.; Müller-Putz, G.R. Directional Decoding From EEG in a Center-Out Motor Imagery Task With Visual and Vibrotactile Guidance. Front. Hum. Neurosci. 2021, 15, 548. [Google Scholar] [CrossRef]
- Ofner, P.; Schwarz, A.; Pereira, J.; Müller-Putz, G.R. Upper limb movements can be decoded from the time-domain of low-frequency EEG. PLoS ONE 2017, 12, e0182578. [Google Scholar] [CrossRef]
- Steyrl, D.; Scherer, R.; Förstner, O.; Müller-Putz, G. Motor Imagery Brain-Computer Interfaces: Random Forests vs Regularized LDA—Non-linear Beats Linear. In Proceedings of the 6th International Brain-Computer Interface Conference Graz 2014, Graz, Austria, 16–19 September 2014; pp. 061-1–061-4. [Google Scholar] [CrossRef]
- Vargic, R.; Chlebo, M.; Kacur, J. Human computer interaction using BCI based on sensorimotor rhythm. In Proceedings of the 2015 IEEE 19th International Conference on Intelligent Engineering Systems (INES), Bratislava, Slovakia, 3–5 September 2015; pp. 91–95. [Google Scholar] [CrossRef]
- Ma, Y.; Ding, X.; She, Q.; Luo, Z.; Potter, T.; Zhang, Y. Classification of motor imagery EEG signals with support vector machines and particle swarm optimization. Comput. Math. Methods Med. 2016, 2016, 4941235. [Google Scholar] [CrossRef]
- Zhang, R.; Xiao, X.; Liu, Z.; Jiang, W.; Li, J.; Cao, Y.; Ren, J.; Jiang, D.; Cui, L. A New Motor Imagery EEG Classification Method FB-TRCSP+RF Based on CSP and Random Forest. IEEE Access 2018, 6, 44944–44950. [Google Scholar] [CrossRef]
- Bentlemsan, M.; Zemouri, E.T.; Bouchaffra, D.; Yahya-Zoubir, B.; Ferroudji, K. Random Forest and Filter Bank Common Spatial Patterns for EEG-Based Motor Imagery Classification. In Proceedings of the 2014 5th International Conference on Intelligent Systems, Modelling and Simulation, Hunan, China, 15–16 June 2014; pp. 235–238. [Google Scholar] [CrossRef]
- Zhang, Z.; Duan, F.; Solé-Casals, J.; Dinarès-Ferran, J.; Cichocki, A.; Yang, Z.; Sun, Z. A Novel Deep Learning Approach With Data Augmentation to Classify Motor Imagery Signals. IEEE Access 2019, 7, 15945–15954. [Google Scholar] [CrossRef]
- Hou, Y.; Jia, S.; Lun, X.; Hao, Z.; Shi, Y.; Li, Y.; Zeng, R.; Lv, J. GCNs-Net: A Graph Convolutional Neural Network Approach for Decoding Time-Resolved EEG Motor Imagery Signals. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Strahnen, M.; Kessler, P. Investigation of a Deep-Learning Based Brain–Computer Interface With Respect to a Continuous Control Application. IEEE Access 2022, 10, 131090–131100. [Google Scholar] [CrossRef]
- Lee, D.Y.; Jeong, J.H.; Lee, B.H.; Lee, S.W. Motor Imagery Classification Using Inter-Task Transfer Learning via a Channel-Wise Variational Autoencoder-Based Convolutional Neural Network. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Tavakolan, M.; Frehlick, Z.; Yong, X.; Menon, C. Classifying three imaginary states of the same upper extremity using time-domain features. PLoS ONE 2017, 12, e0174161. [Google Scholar] [CrossRef]
- Decety, J. The neurophysiological basis of motor imagery. Behav. Brain Res. 1996, 77, 45–52. [Google Scholar] [CrossRef]
- Khademi, S.; Neghabi, M.; Farahi, M.; Shirzadi, M.; Marateb, H.R. A comprehensive review of the movement imaginary brain–computer interface methods: Challenges and future directions. In Artificial Intelligence-Based Brain-Computer Interface; Academic Press: Cambridge, MA, USA, 2022; pp. 23–74. [Google Scholar]
- Battaglia, F.; Quartarone, A.; Ghilardi, M.F.; Dattola, R.; Bagnato, S.; Rizzo, V.; Morgante, L.; Girlanda, P. Unilateral cerebellar stroke disrupts movement preparation and motor imagery. Clin. Neurophysiol. 2006, 117, 1009–1016. [Google Scholar] [CrossRef]
- Müller-Putz, G.R.; Kobler, R.J.; Pereira, J.; Lopes-Dias, C.; Hehenberger, L.; Mondini, V.; Martínez-Cagigal, V.; Srisrisawang, N.; Pulferer, H.; Batistić, L.; et al. Feel your reach: An EEG-based framework to continuously detect goal-directed movements and error processing to gate kinesthetic feedback informed artificial arm control. Front. Hum. Neurosci. 2022, 16, 110. [Google Scholar] [CrossRef]
- Delorme, A.; Makeig, S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef]
- Makeig, S.; Bell, A.J.; Jung, T.P.; Sejnowski, T.J. Independent component analysis of electroencephalographic data. In Advances in Neural Information Processing Systems 8 (NIPS 1995); Touretzky, D., Mozer, M., Hasselmo, M., Eds.; MIT Press: Cambridge, MA, USA, 1996; pp. 145–151. [Google Scholar]
- Chaumon, M.; Bishop, D.V.; Busch, N.A. A practical guide to the selection of independent components of the electroencephalogram for artifact correction. J. Neurosci. Methods 2015, 250, 47–63. [Google Scholar] [CrossRef]
- Balakrishnama, S.; Ganapathiraju, A. Linear Discriminant Analysis—A Brief Tutorial; Technical Report; Institute for Signal and Information Processing: Philadelphia, PA, USA, 1998. [Google Scholar]
- Blankertz, B.; Lemm, S.; Treder, M.; Haufe, S.; Müller, K.R. Single-trial analysis and classification of ERP components—A tutorial. NeuroImage 2011, 56, 814–825. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, MA, USA, 2000. [Google Scholar] [CrossRef]
- Géron, A. Hands-on Machine Learning with Scikit-Learn and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems; O’Reilly Media: Sebastopol, CA, USA, 2017. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Bridle, J.S. Probabilistic Interpretation of Feedforward Classification Network Outputs, with Relationships to Statistical Pattern Recognition. In Proceedings of the NATO Neurocomputing, Les Arcs, France, 27 February–3 March 1989. [Google Scholar]
- Zhou, Y.T.; Chellappa, R. Computation of optical flow using a neural network. In Proceedings of the ICNN, San Diego, CA, USA, 24–27 July 1988; pp. 71–78. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Yang, C.; Kong, L.; Zhang, Z.; Tao, Y.; Chen, X. Exploring the Visual Guidance of Motor Imagery in Sustainable Brain–Computer Interfaces. Sustainability 2022, 14, 13844. [Google Scholar] [CrossRef]
- Krichen, M.; Mihoub, A.; Alzahrani, M.Y.; Adoni, W.Y.H.; Nahhal, T. Are Formal Methods Applicable To Machine Learning Furthermore, Artificial Intelligence? In Proceedings of the 2nd International Conference of Smart Systems and Emerging Technologies (SMARTTECH), Riyadh, Saudi Arabia, 9–11 May 2022; pp. 48–53. [Google Scholar] [CrossRef]
- Raman, R.; Gupta, N.; Jeppu, Y. Framework for Formal Verification of Machine Learning Based Complex System-of-Systems. INSIGHT 2023, 26, 91–102. [Google Scholar] [CrossRef]


| Accuracy (%) | ||||||
|---|---|---|---|---|---|---|
| Classification Method | sLDA | SVM | RF | VGG-19 | ResNet-101 | DenseNet-169 |
| EF vs. EE (0.2–5 Hz) | 53.59 | 53.07 | 53.93 | 57.47 | 72.30 | 66.24 |
| EF vs. EE (1–40 Hz) | 54.75 | 55.47 | 54.03 | 56.24 | 69.82 | 62.94 |
| Method | sLDA | SVM | RF | VGG-19 | ResNet-101 | DenseNet-169 |
|---|---|---|---|---|---|---|
| sLDA | −0.1 | 0.2 | −2.7 | −16.9 * | −10.4 * | |
| SVM | 0.1 | 0.3 | − 2.6 | −16.8 * | −10.3 * | |
| RF | − 0.2 | − 0.3 | − 2.9 | −17.1 * | −10.6 * | |
| VGG-19 | 2.7 | 2.6 | 2.9 | −14.2 * | −7.7 * | |
| ResNet-101 | 16.9 * | 16.8 * | 17.1 * | 14.2 * | 6.5 * | |
| DenseNet-169 | 10.4 * | 10.3 * | 10.6 * | 7.7 * | −6.5 * |
| Classification Method | Cond. | Accuracy (%) | |||||
|---|---|---|---|---|---|---|---|
| sLDA | SVM | RF | VGG-19 | ResNet-101 | DenseNet-169 | ||
| right vs. up (0.2–5 Hz) | VtG | 64.07 | 64.07 | 56.49 | 59.29 | 70.99 | 65.31 |
| noVtG | 60.44 | 59.64 | 55.87 | 60.05 | 70.15 | 65.60 | |
| right vs. up (1–40 Hz) | VtG | 60.87 | 59.38 | 56.96 | 55.63 | 67.93 | 62.13 |
| noVtG | 57.66 | 55.72 | 54.75 | 55.53 | 68.59 | 60.50 | |
| Method | sLDA | SVM | RF | VGG-19 | ResNet-101 | DenseNet-169 |
|---|---|---|---|---|---|---|
| sLDA | 1.0 | 4.7 * | 3.1 | −8.6 * | −2.6 | |
| SVM | −1.0 | 3.7 * | 2.1 | −9.7 * | −3.6 | |
| RF | −4.7 * | −3.7 * | −1.6 | −13.4 * | −7.4 * | |
| VGG-19 | −3.1 | −2.1 | 1.6 | −11.8 * | −5.8 * | |
| ResNet-101 | 8.6 * | 9.7 * | 13.4 * | 11.8 * | 6.0 * | |
| DenseNet-169 | 2.6 | 3.6 | 7.4 * | 5.8 * | −6.0 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batistić, L.; Sušanj, D.; Pinčić, D.; Ljubic, S. Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance. Sensors 2023, 23, 5064. https://doi.org/10.3390/s23115064
Batistić L, Sušanj D, Pinčić D, Ljubic S. Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance. Sensors. 2023; 23(11):5064. https://doi.org/10.3390/s23115064
Chicago/Turabian StyleBatistić, Luka, Diego Sušanj, Domagoj Pinčić, and Sandi Ljubic. 2023. "Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance" Sensors 23, no. 11: 5064. https://doi.org/10.3390/s23115064
APA StyleBatistić, L., Sušanj, D., Pinčić, D., & Ljubic, S. (2023). Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance. Sensors, 23(11), 5064. https://doi.org/10.3390/s23115064







