Classification of Citrus Huanglongbing Degree Based on CBAM-MobileNetV2 and Transfer Learning
Abstract
:1. Introduction
- We created the citrus huanglongbing image dataset ourselves, and based on an improved lightweight deep learning model, we could classify the early, middle, and late stages of citrus huanglongbing leaf images with different degrees of the disease, so as to realize the early identification and classification of citrus huanglongbing.
- We proposed a lightweight deep learning model, CBAM-MobileNetV2, which combined the pretrained MobileNetV2 and an attention module. The convolutional module captured object-based high-level information, while the attention module paid more attention to specific salient regions compared to the convolutional module. Therefore, the convolutional module and attention module delivered complementary information which can more effectively classify the disease severity of citrus huanglongbing.
- Our proposed method requires a smaller number of trainable parameters as we leverage the pre-trained weights for all layers of the MobileNetv2 architecture. This makes our model more suitable for deployment on resource-constrained devices.
- We compared MobileNetV2, InceptionV3, and Xception with the model proposed in this paper. The experimental results showed that CBAM-MobileNetV2 had stable performance on the citrus huanglongbing dataset and outperformed other lightweight models. Furthermore, dropout and data augmentation techniques were incorporated to minimize the chances of overfitting.
2. Materials
2.1. Image Acquisition and Selection
2.2. Image Augmentation
3. Construction of the Classification Model for Citrus Huanglongbing Disease Severity
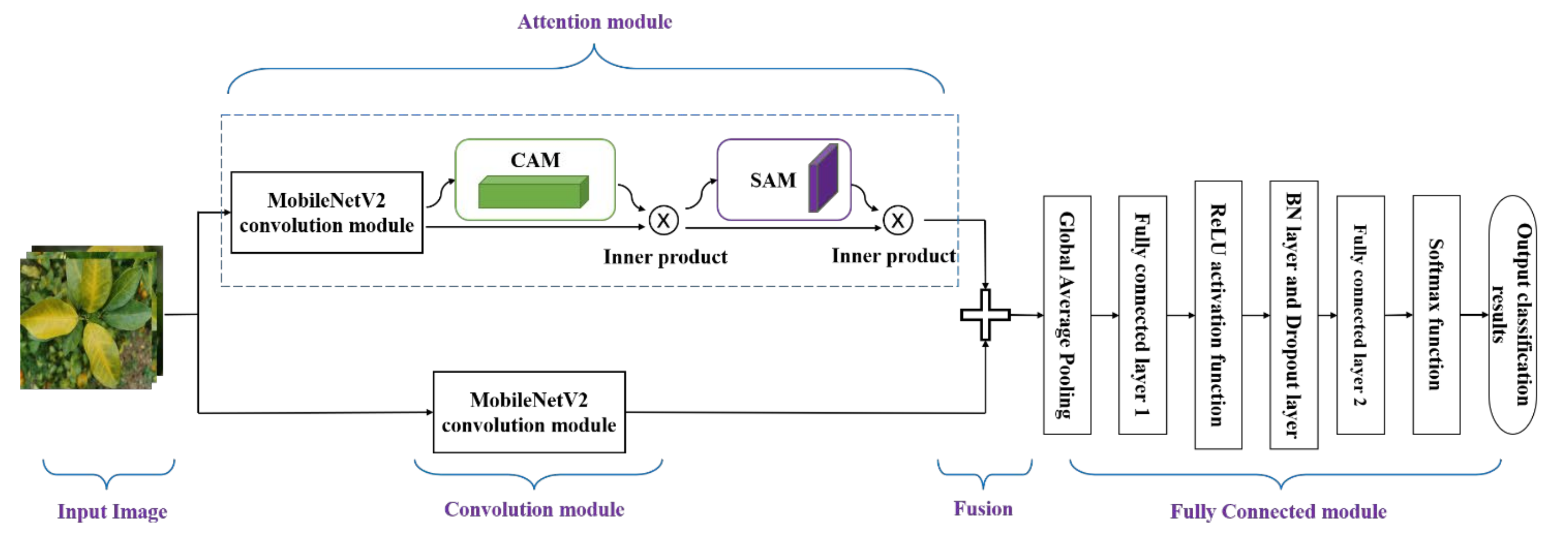
3.1. The Convolution Module
3.2. Convolutional Block Attention Module
3.2.1. Channel Attention Module
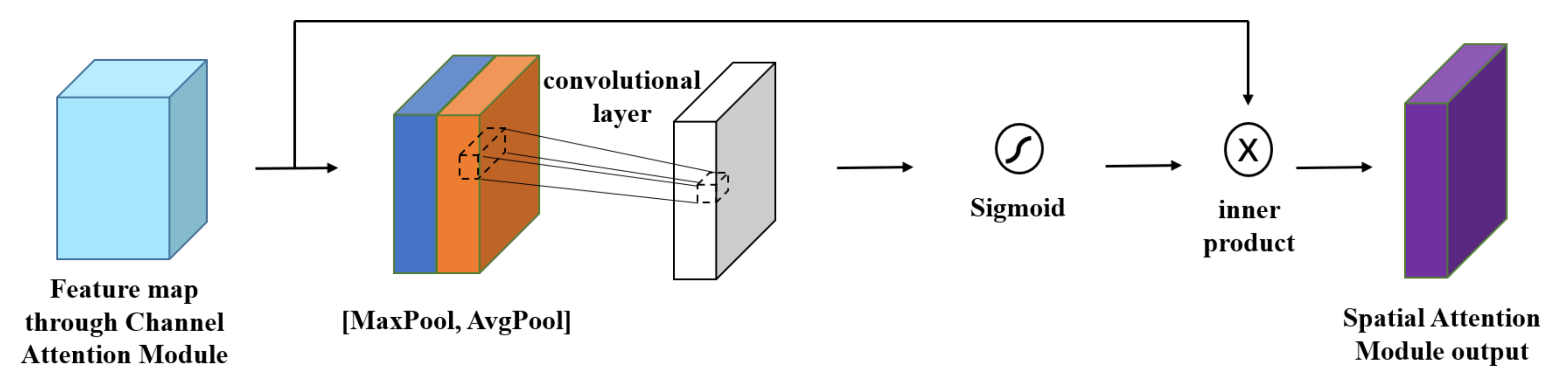
3.2.2. Spatial Attention Module
3.3. ImageNet Dataset
3.4. Transfer Learning
3.4.1. Parameter Fine Tuning
3.4.2. Parameter Freezing
3.5. Model Building and Improvement
3.6. Evaluation Metrics
4. Model Training and Analysis of Experimental Results
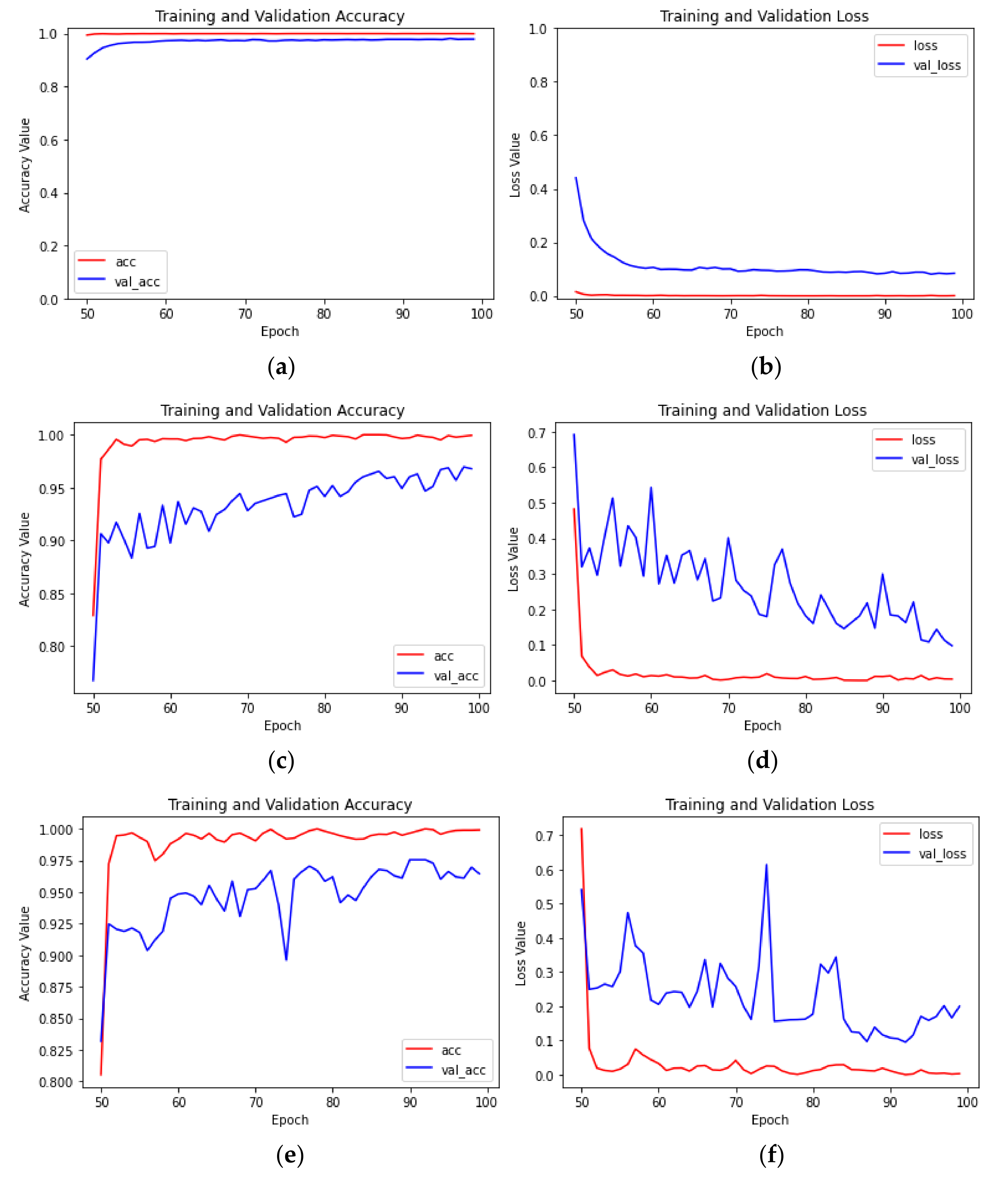
4.1. Experimental Program
4.2. Analysis of Experimental Results
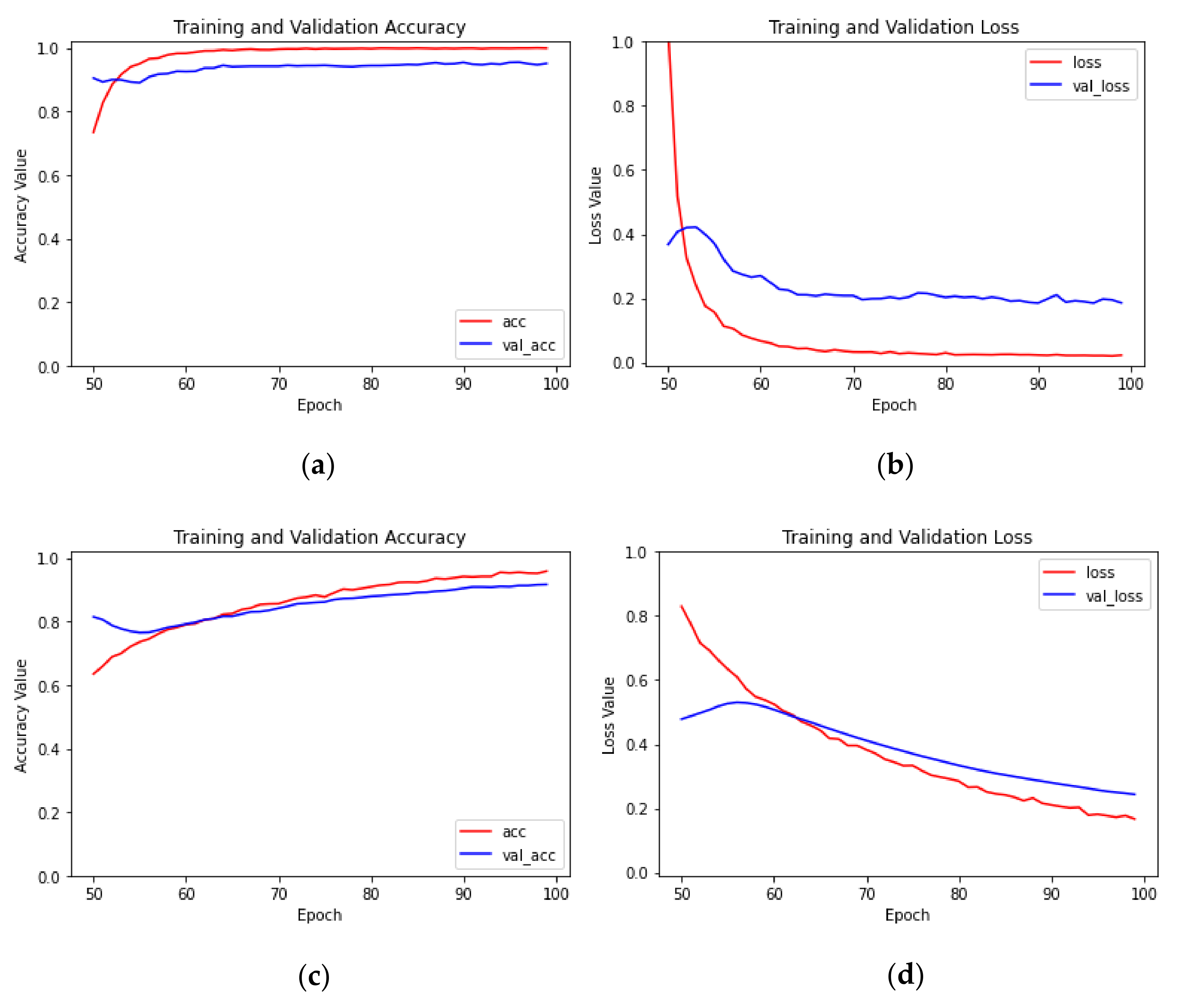
4.2.1. Impact of Transfer Learning Methods on Model Performance
4.2.2. Recognition Performance Analysis of Different Models
4.2.3. Impact of Initial Learning Rates on Model Performance
4.2.4. Comparison of the Latest Classification Methods for Citrus Diseases
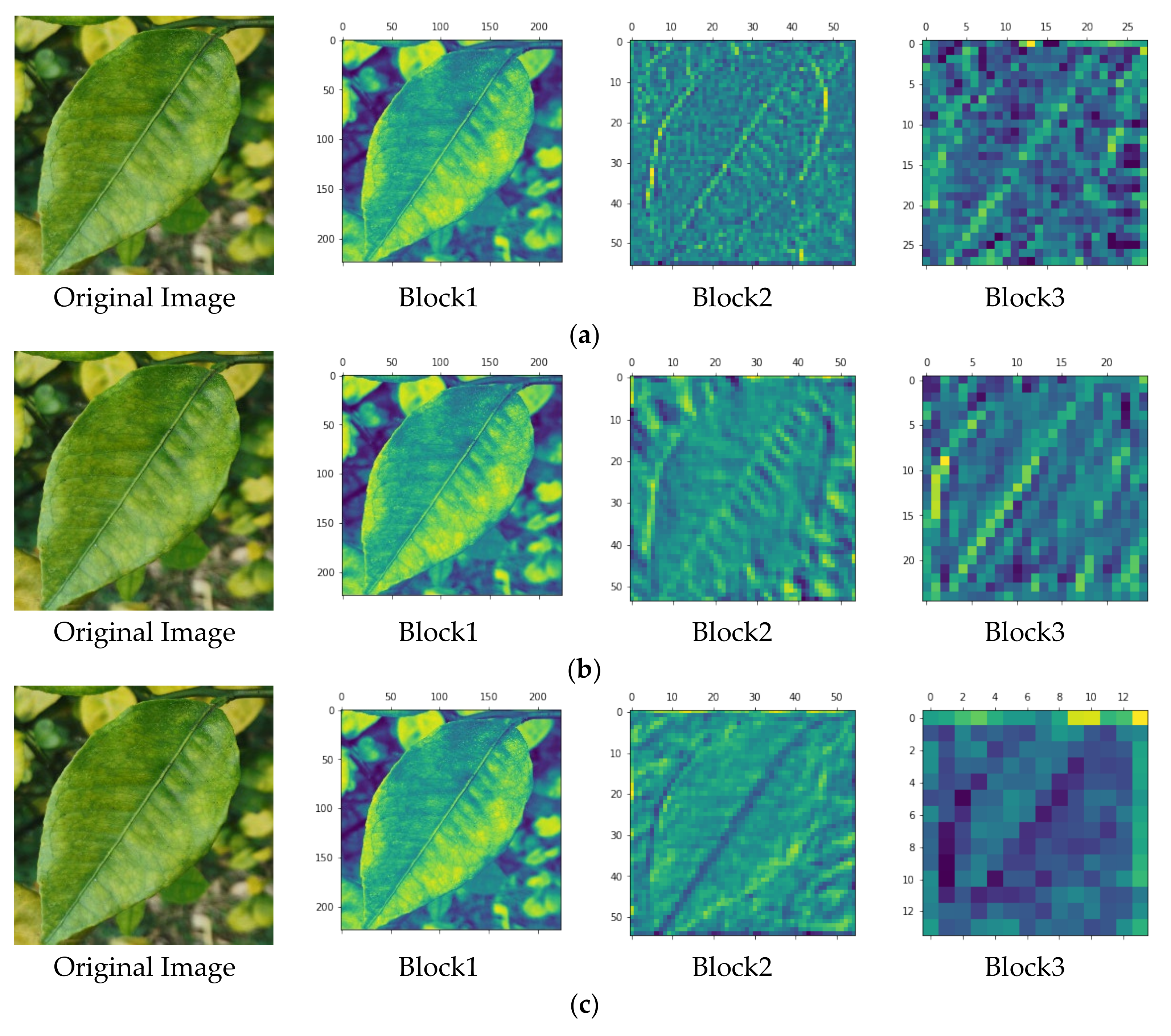
4.3. Ablative Study of the Proposed Method
5. Conclusions
- (1)
- Based on CBAM-MobileNetV2 and transfer learning, the recognition accuracy of the citrus huanglongbing leaf image recognition model of different degrees reached 98.75%, and a very good recognition effect was achieved. The effect was significantly better than that of the MobileNetV2, Xception, and InceptionV3 network models. The convolutional module of CBAM-MobileNetV2 captured object-based high-level information, while the attention module paid more attention to specific salient regions compared to the convolutional module. Therefore, the convolutional module and attention module delivered complementary information which can more effectively classify the disease severity of citrus huanglongbing.
- (2)
- With the same model and initial learning rate, the transfer learning method of parameter fine tuning was significantly better than the method of parameter freezing, and the recognition accuracy of the test set increased by 1.02 to 13.6 percentage points, which showed that the transfer learning method of parameter fine tuning was more suitable for recognizing citrus huanglongbing.
- (3)
- The learning rate was found to have a great impact on the convergence and recognition accuracy of the model. In the transfer learning method of parameter fine tuning, when the learning rate was 0.001, the effect was the best. Therefore, choosing an appropriate learning rate is very important for training the model. In addition, when the collected image samples are preprocessed and data augmentation is completed, the difference in the field shooting scale and shooting angle should be taken into account, and the model performance can be improved by appropriately increasing the sample size.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAM | channel attention mechanism |
| CBAM | convolutional block attention module |
| CNN | convolutional neural network |
| CPU | central process unit |
| DL | deep learning |
| DSC | depth-wise separable convolution |
| GAN | generative adversarial network |
| GPU | graphics processing unit |
| MLP | multilayer perceptron |
| SAM | spatial attention mechanism |
| TL | transfer learning |
References
- Wang, J.; Wu, Y.; Liao, Y.; Chen, Y. Hyperspectral classification of citrus diseased leaves based on convolutional neural network. Inf. Technol. Informatiz. 2020, 3, 84–87. [Google Scholar]
- Liu, Y.; Xiao, H.; Deng, Q.; Zhang, Z.; Sun, X.; Xiao, Y. Nondestructive detection of citrus greening by near infrared spectroscopy. Trans. Chin. Soc. Agric. Eng. 2016, 32, 202–208. [Google Scholar]
- Fan, S.; Ma, W.; Jiang, W.; Zhang, H.; Wang, J.; Li, Q.; He, P.; Peng, L.; Huang, Z. Preliminary study on remote diagnosis technology of citrus Huanglongbing based on deep learning. China Fruits 2022, 4, 76–79+86+133. [Google Scholar]
- Mei, H.; Deng, X.; Hong, T.; Luo, X.; Deng, X. Early detection and grading of citrus huanglongbing using hyperspectral imaging technique. Trans. Chin. Soc. Agric. Eng. 2014, 30, 140–147. [Google Scholar]
- Jia, S.; Yang, D.; Liu, J. Product Image Fine-grained Classification Based on Convolutional Neural Network. J. Shandong Univ. Sci. Technol. (Nat. Sci.) 2014, 33, 91–96. [Google Scholar]
- Jia, S.; Gao, H.; Hang, X. Research Progress on Image Recognition Technology of Crop Pests and Diseases Based on Deep Learning. Trans. Chin. Soc. Agric. Mach. 2019, 50, 313–317. [Google Scholar]
- Jeon, W.S.; Rhee, S.Y. Plant leaf recognition using a convolution neural network. Int. J. Fuzzy Log. Intell. Syst. 2017, 17, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A convolution neural network-based seed classification system. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep learning for tomato diseases: Classification and symptoms visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, Z.; Dong, W.; Zhang, L.; Rao, Y.; Li, S. Image recognition of tea plant disease based on convolutional neural network and small samples. Jiangsu J. Agric. Sci. 2019, 35, 48–55. [Google Scholar]
- Zhang, J.; Kong, F.; Wu, J.; Zhai, Z.; Han, S.; Cao, S. Cotton disease identification model based on improved VGG convolution neural network. J. China Agric. Univ. 2018, 23, 161–171. [Google Scholar]
- Long, M.; Ouyang, C.; Liu, H.; Fu, Q. Image recognition of Camellia oleifera diseases based on convolutional neural network and transfer learning. Trans. Chin. Soc. Agric. Eng. 2018, 34, 194–201. [Google Scholar]
- Feng, X.; Li, D.; Wang, W.; Zheng, G.; Liu, H.; Sun, Y.; Liang, S.; Yang, Y.; Zang, H.; Zhang, H. Image Recognition of Wheat Leaf Diseases Based on Lightweight Convolutional Neural Network and Transfer Learning. J. Henan Agric. Sci. 2021, 50, 174–180. [Google Scholar]
- Li, S.; Chen, C.; Zhu, T.; Liu, B. Plant Leaf Disease Identification Based on Lightweight Residual Network. Trans. Chin. Soc. Agric. Mach. 2022, 53, 243–250. [Google Scholar]
- Su, S.; Qiao, Y.; Rao, Y. Recognition of grape leaf diseases and mobile application based on transfer learning. Trans. Chin. Soc. Agric. Eng. 2021, 37, 127–134. [Google Scholar]
- Zheng, Y.; Zhang, L. Plant Leaf Image Recognition Method Based on Transfer Learning with Convolutional Neural Networks. Trans. Chin. Soc. Agric. Mach. 2018, 49, 354–359. [Google Scholar]
- Li, M.; Wang, J.; Li, H.; Hu, Z.; Yang, X.; Huang, X.; Zeng, W.; Zhang, J.; Fang, S. Method for identifying crop disease based on CNN and transfer learning. Smart Agric. 2019, 1, 46–55. [Google Scholar]
- Chen, G.; Zhao, S.; Cao, L.; Fu, S.; Zhou, J. Corn plant disease recognition based on migration learning and convolutional neural network. Smart Agric. 2019, 1, 34–44. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted residuals and linear bottleneck. In Proceedings of the 2018 IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- da Silva, J.C.F.; Silva, M.C.; Luz, E.J.S.; Delabrida, S.; Oliveira, R.A.R. Using Mobile Edge AI to Detect and Map Diseases in Citrus Orchards. Sensors 2023, 23, 2165. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, D.; Xi, L. The research of maize disease identification based on MobileNetV2 and transfer learning. J. Henan Agric. Univ. 2022, 56, 1041–1051. [Google Scholar]
- Gulzar, Y. Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability 2023, 15, 1906. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, W. Grape ripeness discrimination based on MobileNetV2. Xinjiang Agric. Mech. 2022, 216, 29–31+42. [Google Scholar]
- Yang, M.; Zhang, Y.; Liu, T. Corn disease recognition based on the Convolutional Neural Network with a small sampling size. Chin. J. Eco-Agric. 2020, 28, 1924–1931. [Google Scholar]
- Xiang, Q.; Wang, X.; Li, R.; Zhang, G.; Lai, J.; Hu, Q. Fruit image classification based on Mobilenetv2 with transfer learning technique. In Proceedings of the 3rd International Conference on Computer Science and Application Engineering, Sanya, China, 22–24 October 2019; pp. 1–7. [Google Scholar]
- Chen, J.; Zhang, D.; Nanehkaran, Y. Identifying plant diseases using deep transfer learning and enhanced lightweight network. Multimed. Tools Appl. 2020, 79, 31497–31515. [Google Scholar] [CrossRef]
- Hossain, S.M.M.; Deb, K.; Dhar, P.K.; Koshiba, T. Plant leaf disease recognition using depth-wise separable convolution-based models. Symmetry 2021, 13, 511. [Google Scholar] [CrossRef]
- Liu, G.; Peng, J.; El-Latif, A.A.A. SK-MobileNet: A Lightweight Adaptive Network Based on Complex Deep Transfer Learning for Plant Disease Recognition. Arab. J. Sci. Eng. 2023, 48, 1661–1675. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Sitaula, C.; Xiang, Y.; Aryal, S.; Lu, X. Scene image representation by foreground, background and hybrid features. Expert Syst. Appl. 2021, 182, 115285. [Google Scholar] [CrossRef]
- Mishra, B.; Shahi, T.B. Deep learning-based framework for spatiotemporal data fusion: An instance of Landsat 8 and Sentinel 2 NDVI. J. Appl. Remote Sens. 2021, 15, 034520. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the 2018 European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 3–9. [Google Scholar]
- Deng, X.; Lan, Y.; Hong, T.; Chen, J. Citrus greening detection using visible spectrum imaging and C-SVC. Comput. Electron. Agric. 2016, 130, 177–183. [Google Scholar] [CrossRef]
- Sharif, M.; Khan, M.A.; Iqbal, Z.; Azam, M.F.; Lali, M.I.U.; Javed, M.Y. Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Comput. Electron. Agric. 2018, 150, 220–234. [Google Scholar] [CrossRef]
- Pan, W.; Qin, J.; Xiang, X.; Wu, Y.; Tan, Y.; Xiang, L. A smart mobile diagnosis system for citrus diseases based on densely connected convolutional networks. IEEE Access 2019, 7, 87534–87542. [Google Scholar] [CrossRef]
- Xing, S.; Lee, M.; Lee, K. Citrus pests and diseases recognition model using weakly dense connected convolution network. Sensors 2019, 19, 3195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tie, J.; Luo, J.; Zheng, L.; Mo, H.; Long, J. Citrus disease recognition based on improved residual network. J. South-Cent. Minzu Univ. (Nat. Sci. Ed.) 2021, 40, 621–630. [Google Scholar]




| Degrees of Disease | Number of Original Images | Number of Images after Augmentation | Number of Images | |
|---|---|---|---|---|
| Training Set | Test Set | |||
| Early stage | 295 | 2360 | 1888 | 472 |
| Middle stage | 253 | 2024 | 1619 | 405 |
| Late stage | 203 | 1624 | 1299 | 325 |
| Number of Layers | Input Size | Operator and Convolution Kernel | N | S |
|---|---|---|---|---|
| 1 | 224 × 224 × 3 | Conv2d 3 × 3 | 1 | 2 |
| 2 | 112 × 112 × 32 | Bottleneck 3 × 3 1 × 1 | 1 | 1 |
| 3–4 | 112 × 112 × 16 | Bottleneck 3 × 3 1 × 1 | 2 | 2 |
| 5–7 | 56 × 56 × 24 | Bottleneck 3 × 3 1 × 1 | 3 | 2 |
| 8–11 | 28 × 28 × 32 | Bottleneck 3 × 3 1 × 1 | 4 | 2 |
| 12–14 | 14 × 14 × 64 | Bottleneck 3 × 3 1 × 1 | 3 | 1 |
| 15–17 | 14 × 14 × 96 | Bottleneck 3 × 3 1 × 1 | 3 | 2 |
| 18 | 7 × 7 × 160 | Bottleneck 3 × 3 1 × 1 | 1 | 1 |
| 19 | 7 × 7 × 320 | Conv2d 1 × 1 | 1 | 1 |
| 20 | 7 × 7 × 1280 | Avgpool 7 × 7 | 1 | - |
| 21 | 1 × 1 × 1280 | Conv2d 1 × 1 | 1 | - |
| Scheme No. | Transfer Learning Method | Model | Initial Learning Rate | Number of Parameters | Training Time | Recognition Accuracy | |
|---|---|---|---|---|---|---|---|
| Training Set | Test Set | ||||||
| 1 | Parameter freezing | CBAM-MobileNetV2 | 0.001 | 453,574 | 419.01 m | 99.83 | 93.50 |
| 2 | 0.0001 | 425.21 m | 100.00 | 92.31 | |||
| 3 | 0.00001 | 423.54 m | 95.40 | 89.70 | |||
| 4 | MobileNetV2 | 0.001 | 329,219 | 413.02 m | 99.60 | 93.16 | |
| 5 | 0.0001 | 421.03 m | 99.92 | 92.15 | |||
| 6 | 0.00001 | 425.93 m | 86.90 | 83.11 | |||
| 7 | Xception | 0.001 | 525,827 | 765.95 m | 98.80 | 86.74 | |
| 8 | 0.0001 | 772.00 m | 99.63 | 90.03 | |||
| 9 | 0.00001 | 765.48 m | 78.81 | 75.59 | |||
| 10 | InceptionV3 | 0.001 | 525,827 | 531.60 m | 99.12 | 88.34 | |
| 11 | 0.0001 | 520.22 m | 99.52 | 88.68 | |||
| 12 | 0.00001 | 501.13 m | 80.37 | 75.93 | |||
| 13 | Parameter Fine tuning | CBAM-MobileNetV2 | 0.001 | 2,371,462 | 480.25 m | 100.00 | 98.75 |
| 14 | 0.0001 | 476.05 m | 100.00 | 95.52 | |||
| 15 | 0.00001 | 473.87 m | 95.85 | 92.48 | |||
| 16 | MobileNetV2 | 0.001 | 2,191,811 | 472.38 m | 100.00 | 98.14 | |
| 17 | 0.0001 | 478.96 m | 100.00 | 94.85 | |||
| 18 | 0.00001 | 466.62 m | 95.31 | 92.15 | |||
| 19 | Xception | 0.001 | 10,004,171 | 964.67 m | 100.00 | 96.96 | |
| 20 | 0.0001 | 982.75 m | 100.00 | 91.05 | |||
| 21 | 0.00001 | 973.13 m | 92.98 | 87.58 | |||
| 22 | InceptionV3 | 0.001 | 14,149,699 | 633.37 m | 100.00 | 97.55 | |
| 23 | 0.0001 | 596.86 m | 100.00 | 91.30 | |||
| 24 | 0.00001 | 603.23 m | 99.75 | 89.53 | |||
| Classification Method | Dataset | Classes | Availability | Accuracy |
|---|---|---|---|---|
| C-SVC [34] | Self-created [34] | 2 | Private | 91.93% |
| M-SVM [35] | Citrus disease image gallery [35] | 6 | Public | 97.00% |
| Simplify DenseNet201 [36] | Self-created [36] | 6 | Private | 88.77% |
| Weakly DenseNet-16 [37] | Self-created [37] | 24 | Public | 93.33% |
| F-ResNet [38] | PlantVillage and self-created [38] | 5 | Private | 93.60% |
| CBAM-MobileNetV2 | Self-created | 3 | Private | 98.75% |
| Transfer Learning Method | Initial Learning Rate | Attention (with or without) | Recognition Accuracy | |
|---|---|---|---|---|
| Training Set | Test Set | |||
| Parameter fine tuning | 0.001 | Attention (with) | 100.00 | 98.75 |
| Attention (without) | 100.00 | 98.14 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, S.; Wang, L.; Fan, D.; Miao, L.; Yan, J.; He, H. Classification of Citrus Huanglongbing Degree Based on CBAM-MobileNetV2 and Transfer Learning. Sensors 2023, 23, 5587. https://doi.org/10.3390/s23125587
Dou S, Wang L, Fan D, Miao L, Yan J, He H. Classification of Citrus Huanglongbing Degree Based on CBAM-MobileNetV2 and Transfer Learning. Sensors. 2023; 23(12):5587. https://doi.org/10.3390/s23125587
Chicago/Turabian StyleDou, Shiqing, Lin Wang, Donglin Fan, Linlin Miao, Jichi Yan, and Hongchang He. 2023. "Classification of Citrus Huanglongbing Degree Based on CBAM-MobileNetV2 and Transfer Learning" Sensors 23, no. 12: 5587. https://doi.org/10.3390/s23125587
APA StyleDou, S., Wang, L., Fan, D., Miao, L., Yan, J., & He, H. (2023). Classification of Citrus Huanglongbing Degree Based on CBAM-MobileNetV2 and Transfer Learning. Sensors, 23(12), 5587. https://doi.org/10.3390/s23125587




