Detection of Colorectal Polyps from Colonoscopy Using Machine Learning: A Survey on Modern Techniques
Abstract
:1. Introduction
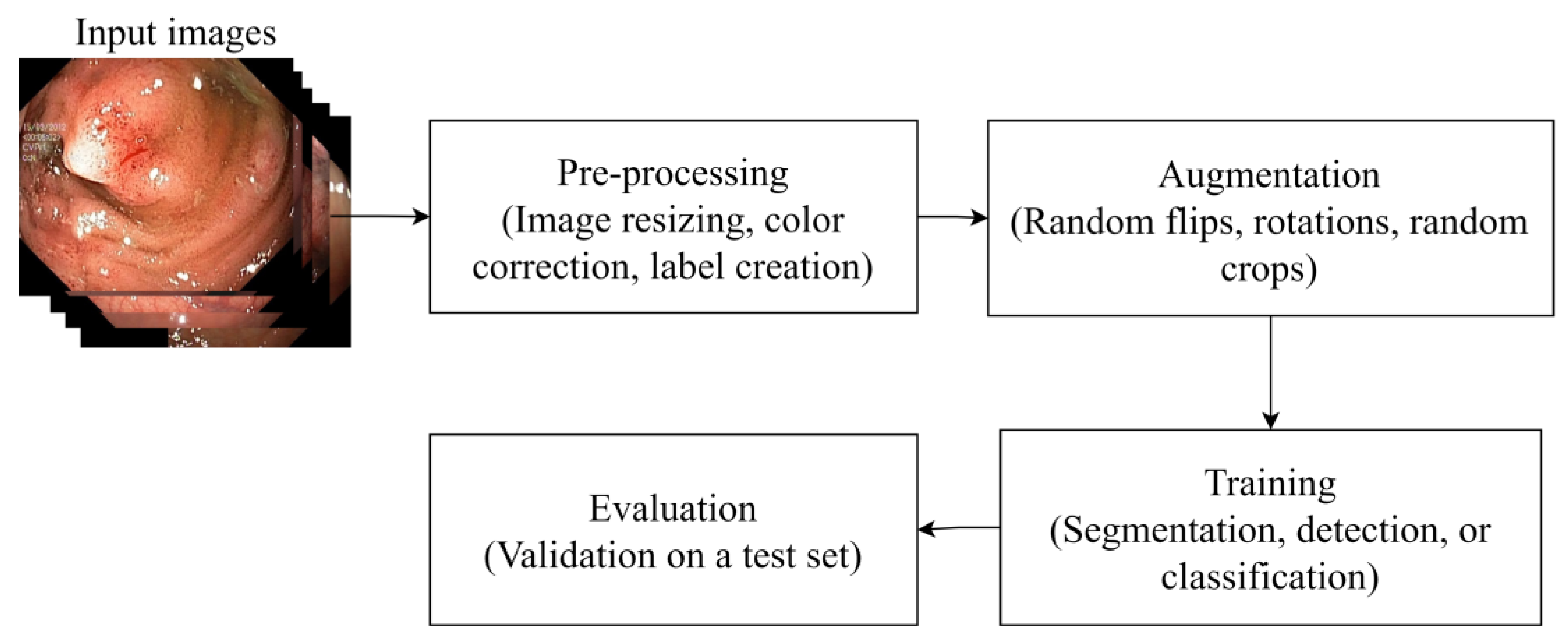
2. Survey Strategy
3. Contributions
- We present a review of several recently proposed colorectal polyp detection methods.
- We review and analyze the various architectures available to build colorectal polyp detectors.
- Based on the current work, we present several common challenges researchers face when building colorectal polyp detection algorithms.
- We present an analysis of the existing benchmark colorectal polyps datasets.
- We analyze the findings in the reviewed literature and investigate the current gaps and trends.
4. Common Challenges
4.1. Data Disparity
4.2. Poor Bowel Preparation
4.3. High Demand for Computational Resources
4.4. Colonoscopy Light Reflection
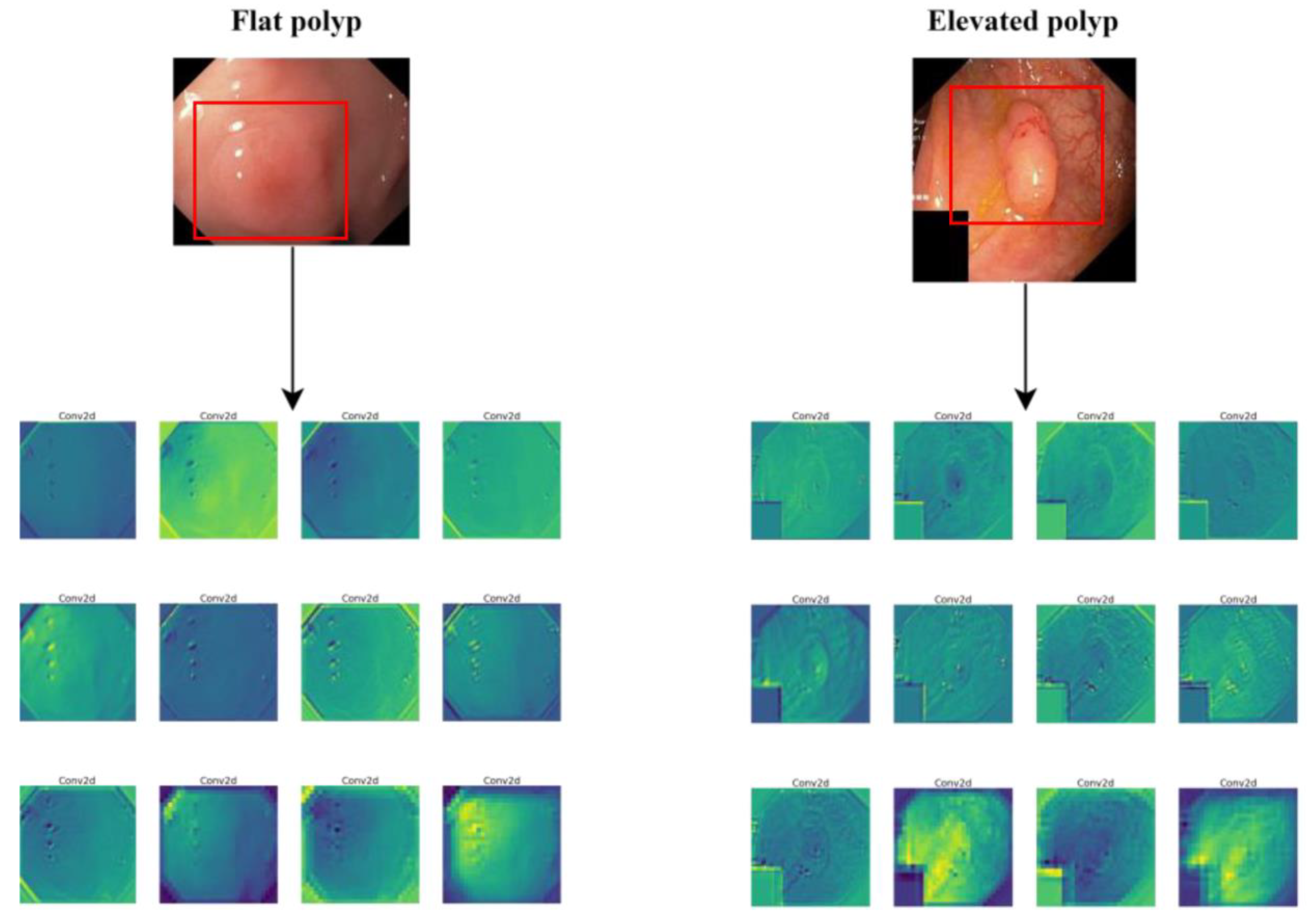
4.5. Colonoscopy Viewpoints
4.6. Pre-Training on Samples from an Irrelevant Domain
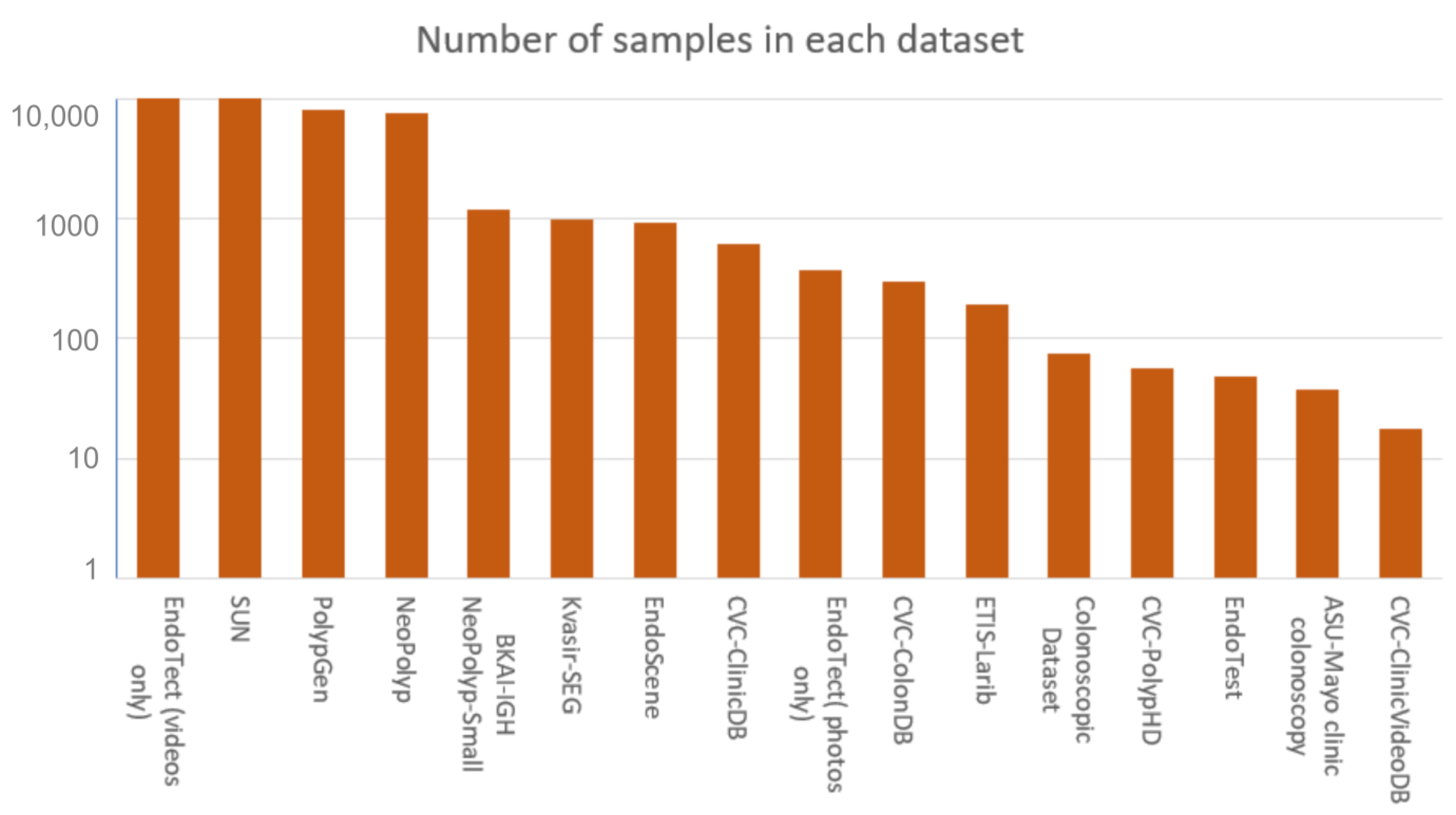
5. Benchmark Datasets
5.1. Kvasir-SEG
5.2. ETIS-Larib
5.3. CVC-ClinicDB
5.4. CVC-ColonDB
5.5. CVC-PolypHD
5.6. EndoTect
5.7. BKAI-IGH NeoPolyp-Small
5.8. NeoPolyp
5.9. PolypGen
5.10. EndoScene
5.11. S.U.N. Colonoscopy Video Database
5.12. EndoTest
5.13. ASU-Mayo Clinic Colonoscopy
5.14. Colonoscopic Dataset
5.15. CVC-ClinicVideoDB
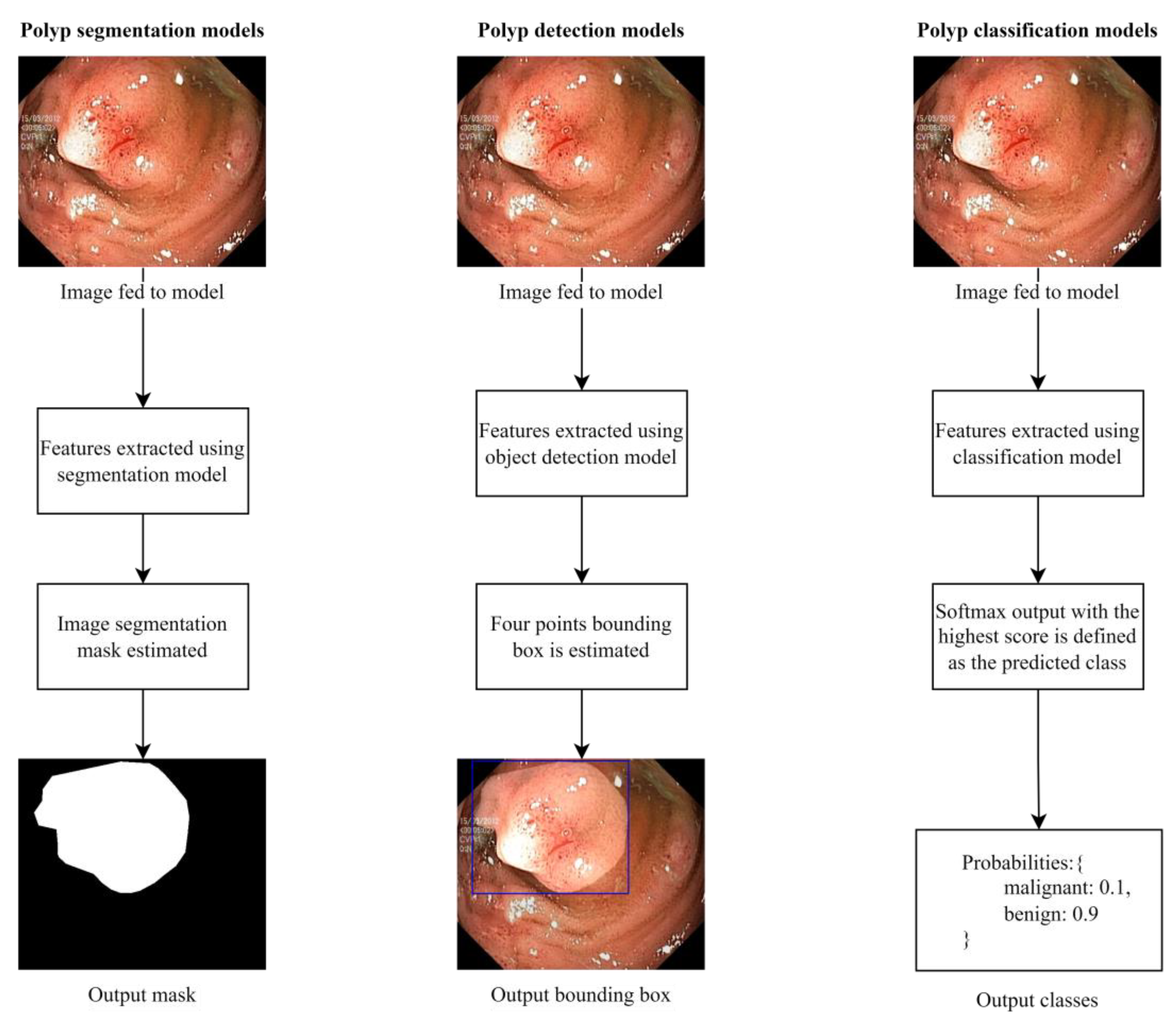
6. Common Architectures
6.1. Segmentation Methods
6.1.1. U-NET
6.1.2. SegNet
6.1.3. Fully Convolutional Networks (FCN)
6.1.4. Pyramid Scene Parsing Network (PSPNet)
6.2. Object Detection Methods
6.2.1. Region-Based Convolutional Neural Networks (RCNN)
6.2.2. Faster Region-Based Convolutional Neural Networks (Faster R-CNN)
6.2.3. Single-Shot Detector (SSD)
6.2.4. You Only Look Once (YOLO)
6.3. Pre-Trained Convolutional Neural Network Models
6.3.1. VGG16
6.3.2. VGG19
6.3.3. ResNet50
6.3.4. Xception
6.3.5. AlexNet
6.3.6. GoogLeNet
7. Performance Evaluation
7.1. Classification and Localization Metrics
7.2. Segmentation Metrics
8. Survey of Existing Work
9. Discussion and Recommendations
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bond, J.H. Polyp guideline: Diagnosis, treatment, and surveillance for patients with colorectal polyps. Am. J. Gastroenterol. 2000, 95, 3053–3063. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wang, Y.; Qi, M.; He, X.; Zhu, Y.; Hong, J. Risk factors for recurrent colorectal polyps. Gut Liver 2020, 14, 399–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shussman, N.; Wexner, S.D. Colorectal polyps and polyposis syndromes. Gastroenterol. Rep. 2014, 2, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Colorectal Cancer. 2022. Available online: https://www.iarc.who.int/cancer-type/colorectal-cancer/ (accessed on 20 December 2022).
- Le Marchand, L.; Wilkens, L.R.; Kolonel, L.N.; Hankin, J.H.; Lyu, L.C. Associations of sedentary lifestyle, obesity, smoking, alcohol use, and diabetes with the risk of colorectal cancer. Cancer Res. 1997, 57, 4787–4794. [Google Scholar] [PubMed]
- Jeong, Y.H.; Kim, K.O.; Park, C.S.; Kim, S.B.; Lee, S.H.; Jang, B.I. Risk factors of advanced adenoma in small and diminutive colorectal polyp. J. Korean Med. Sci. 2016, 31, 1426–1430. [Google Scholar] [CrossRef]
- Williams, C.; Teague, R. Progress report colonoscopy. Gut 1973, 14, 990–1003. [Google Scholar] [CrossRef] [Green Version]
- Pacal, I.; Karaman, A.; Karaboga, D.; Akay, B.; Basturk, A.; Nalbantoglu, U.; Coskun, S. An efficient real-time colonic polyp detection with YOLO algorithms trained by using negative samples and large datasets. Comput. Biol. Med. 2022, 141, 105031. [Google Scholar] [CrossRef]
- Nisha, J.; Gopi, V.P.; Palanisamy, P. Automated colorectal polyp detection based on image enhancement and dual-path CNN architecture. Biomed. Signal. Process Control 2022, 73, 103465. [Google Scholar] [CrossRef]
- Yang, K.; Chang, S.; Tian, Z.; Gao, C.; Du, Y.; Zhang, X.; Liu, K.; Meng, J.; Xue, L. Automatic polyp detection and segmentation using shuffle efficient channel attention network. Alex. Eng. J. 2022, 61, 917–926. [Google Scholar] [CrossRef]
- Gong, E.J.; Bang, C.S.; Lee, J.J.; Seo, S.I.; Yang, Y.J.; Baik, G.H.; Kim, J.W. No-Code Platform-Based Deep-Learning Models for Prediction of Colorectal Polyp Histology from White-Light Endoscopy Images: Development and Performance Verification. J. Pers. Med. 2022, 12, 963. [Google Scholar] [CrossRef]
- Puyal, J.G.-B.; Brandao, P.; Ahmad, O.F.; Bhatia, K.K.; Toth, D.; Kader, R.; Lovat, L.; Mountney, P.; Stoyanov, D. Polyp detection on video colonoscopy using a hybrid 2D/3D CNN. Med. Image Anal. 2022, 82, 102625. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Zhao, L.; Feng, S.; Zhang, S.; Zhou, Q.; Gao, X.; Guo, Y. Colorectal polyp region extraction using saliency detection network with neutrosophic enhancement. Comput. Biol. Med. 2022, 147, 105760. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.K.; Yildirim-Yayilgan, S.; Farup, I.; Pedersen, M.; Hovde, O. Y-Net: A deep Convolutional Neural Network to Polyp Detection. In Proceedings of the British Machine Vision Conference 2018, BMVC 2018, Tyne, UK, 3–6 September 2018; pp. 1–11. [Google Scholar]
- Umehara, K.; Näppi, J.J.; Hironaka, T.; Regge, D.; Ishida, T.; Yoshida, H. Medical Imaging: Computer-Aided Diagnosis—Deep ensemble learning of virtual endoluminal views for polyp detection in CT colonography. SPIE Proc. 2017, 10134, 108–113. [Google Scholar]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; Campos-Tato, F.; Herrero, J.; Puga, M.; Remedios, D.; Rivas, L.; Sánchez, E.; Iglesias, A.; Cubiella, J.; et al. Real-time polyp detection model using convolutional neural networks. Neural Comput. Appl. 2022, 34, 10375–10396. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; Lange, T.D.; Johansen, D.; Johansen, H.D. Kvasir-SEG: A Segmented Polyp Dataset. In International Conference on Multimedia Modeling; Springer: Berlin, Germany, 2020; pp. 451–462. [Google Scholar] [CrossRef]
- Silva, J.S.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef]
- Haj-Manouchehri, A.; Mohammadi, H.M. Polyp detection using CNNs in colonoscopy video. IET Comput. Vis. 2020, 14, 241–247. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; de Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A Comprehensive Study on Colorectal Polyp Segmentation with ResUNet++, Conditional Random Field and Test-Time Augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Available online: http://lmb.informatik.uni-freiburg.de/ (accessed on 21 December 2022).
- Bernal, J.; Sánchez, J.; Vilariño, F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012, 45, 3166–3182. [Google Scholar] [CrossRef]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A Benchmark for Endoluminal Scene Segmentation of Colonoscopy Images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef]
- International Conference on Pattern Recognition, EndoTect 2020. 2020. Available online: https://endotect.com/ (accessed on 21 December 2022).
- An, N.S.; Lan, P.N.; Hang, D.V.; Long, D.V.; Trung, T.Q.; Thuy, N.T.; Sang, D.V. BlazeNeo: Blazing Fast Polyp Segmentation and Neoplasm Detection. IEEE Access. 2022, 10, 43669–43684. [Google Scholar] [CrossRef]
- Lan, P.N.; An, N.S.; Hang, D.V.; Van Long, D.; Trung, T.Q.; Thuy, N.T.; Sang, D.V. NeoUNet: Towards Accurate Colon Polyp Segmentation and Neoplasm Detection. In Advances in Visual Computing; ISVC 2021. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 13018. [Google Scholar] [CrossRef]
- Ali, S.; Jha, D.; Ghatwary, N.; Realdon, S.; Cannizzaro, R.; Salem, O.E.; Lamarque, D.; Daul, C.; Riegler, M.A.; Anonsen, K.V.; et al. PolypGen: A multi-center polyp detection and segmentation dataset for generalisability assessment. arXiv 2021, arXiv:2106.04463. [Google Scholar]
- Ali, S.; Ghatwary, N.; Jha, D.; Isik-Polat, E.; Polat, G.; Yang, C.; Li, W.; Galdran, A.; Ballester, M.-Á.G.; Thambawita, V.; et al. East, Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge. arXiv 2022, arXiv:2202.12031. [Google Scholar]
- SAli, S.; Dmitrieva, M.; Ghatwary, N.; Bano, S.; Polat, G.; Temizel, A.; Krenzer, A.; Hekalo, A.; Guo, Y.B.; Matuszewski, B.; et al. Deep learning for detection and segmentation of artefact and disease instances in gastrointestinal endoscopy. Med. Image Anal. 2021, 70, 102002. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Hotta, K.; Ohtsuka, K.; Matsuda, T.; Saito, S.; Kudo, T.; Baba, T.; Ishida, F.; et al. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointest. Endosc. 2021, 93, 960–967.e3. [Google Scholar] [CrossRef]
- Fitting, D.; Krenzer, A.; Troya, J.; Banck, M.; Sudarevic, B.; Brand, M.; Böck, W.; Zoller, W.G.; Rösch, T.; Puppe, F.; et al. A video based benchmark data set (ENDOTEST) to evaluate computer-aided polyp detection systems. Scand. J. Gastroenterol. 2022, 57, 1397–1403. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated Polyp Detection in Colonoscopy Videos Using Shape and Context Information. IEEE Trans. Med. Imaging. 2016, 35, 630–644. [Google Scholar] [CrossRef]
- Mesejo, P.; Pizarro, D.; Abergel, A.; Rouquette, O.; Beorchia, S.; Poincloux, L.; Bartoli, A. Computer-Aided Classification of Gastrointestinal Lesions in Regular Colonoscopy. IEEE Trans. Med. Imaging. 2016, 35, 2051–2063. [Google Scholar] [CrossRef] [Green Version]
- Angermann, Q.; Bernal, J.; Sánchez-Montes, C.; Hammami, M.; Fernández-Esparrach, G.; Dray, X.; Romain, O.; Sánchez, F.J.; Histace, A. Towards real-time polyp detection in colonoscopy videos: Adapting still frame-based methodologies for video sequences analysis. In Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures; Cardoso, M.J., Arbel, T., Luo, X., Wesarg, S., Reichl, T., González Ballester, M.Á., McLeod, J., Drechsler, K., Peters, T., Erdt, M., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 29–41. [Google Scholar]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Safarov, S.; Whangbo, T. A-DenseUNet: Adaptive Densely Connected UNet for Polyp Segmentation in Colonoscopy Images with Atrous Convolution. Sensors 2021, 21, 1441. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, P.; Wang, D.; Cao, Y.; Liu, B. Colorectal polyp segmentation by U-Net with dilation convolution. In Proceedings of the 18th IEEE International Conference on Machine Learning and Applications. ICMLA, Boca Raton, FL, USA, 16–19 December 2019; Volume 2019, pp. 851–858. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 2015. Available online: http://www.robots.ox.ac.uk/ (accessed on 22 December 2022).
- Afify, H.M.; Mohammed, K.K.; Hassanien, A.E. An improved framework for polyp image segmentation based on SegNet architecture. Int. J. Imaging Syst. Technol. 2021, 31, 1741–1751. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J. Biomed. Health Inform. 2017, 21, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Brandao, P.; Mazomenos, E.; Ciuti, G.; Caliò, R.; Bianchi, F.; Menciassi, A.; Dario, P.; Koulaouzidis, A.; Arezzo, A.; Stoyanov, D. Fully convolutional neural networks for polyp segmentation in colonoscopy. Med. Imaging 2017 Comput.-Aided Diagn. 2017, 10134, 101–107. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Seal, A.; Ojha, A. Localization of Polyps in WCE Images Using Deep Learning Segmentation Methods: A Comparative Study. Commun. Comput. Inf. Sci. CCIS 2022, 1567, 538–549. [Google Scholar] [CrossRef]
- Yadav, N.; Binay, U. Comparative study of object detection algorithms. Int. Res. J. Eng. Technol. 2017, 4, 586–591. Available online: http://www.irjet.net (accessed on 23 December 2022).
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar] [CrossRef] [Green Version]
- Tashk, A.; Nadimi, E. An Innovative Polyp Detection Method from Colon Capsule Endoscopy Images Based on A Novel Combination of RCNN and DRLSE. In Proceedings of the 2020 IEEE Congress on Evolutionary Computation (CEC), Glasgow, UK, 19–24 July 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Qadir, H.A.; Shin, Y.; Solhusvik, J.; Bergsland, J.; Aabakken, L.; Balasingham, I. Polyp Detection and Segmentation using Mask R-CNN: Does a Deeper Feature Extractor CNN Always Perform Better? In Proceedings of the 2019 13th International Symposium on Medical Information and Communication Technology (ISMICT), Oslo, Norway, 8–10 May 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Chen, B.-L.; Wan, J.-J.; Chen, T.-Y.; Yu, Y.-T.; Ji, M. A self-attention based faster R-CNN for polyp detection from colonoscopy images. Biomed. Signal. Process Control. 2021, 70, 103019. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. Available online: https://github.com/ (accessed on 23 December 2022).
- Qian, Z.; Lv, Y.; Lv, D.; Gu, H.; Wang, K.; Zhang, W.; Gupta, M.M. A new approach to polyp detection by pre-processing of images and enhanced faster r-cnn. IEEE Sens. J. 2021, 21, 11374–11381. [Google Scholar] [CrossRef]
- Nadimi, E.S.; Buijs, M.M.; Herp, J.; Kroijer, R.; Kobaek-Larsen, M.; Nielsen, E.; Pedersen, C.D.; Blanes-Vidal, V.; Baatrup, G. Application of deep learning for autonomous detection and localization of colorectal polyps in wireless colon capsule endoscopy. Comput. Electr. Eng. 2020, 81, 106531. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Chang, D.; Hu, Y. Computer-assisted detection of colonic polyps using improved faster R-CNN. Chin. J. Electron. 2019, 28, 718–724. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Computer Vision—ECCV 2016; Lecture Notes in Computer Science; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Cham, Switzerland, 2016; Volume 9905. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Jiang, J.; Wang, Z. Colonic Polyp Detection in Endoscopic Videos with Single Shot Detection Based Deep Convolutional Neural Network. IEEE Access. 2019, 7, 45058–45066. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, S.; Vijayalakshmi, S.; Sabharwal, M.; Kaur, M.; AlZubi, A.A.; Lee, H.-N. Detection and Classification of Colorectal Polyp Using Deep Learning. Biomed. Res. Int. 2022, 2022, 2805607. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. Available online: http://pjreddie.com/yolo/ (accessed on 23 December 2022).
- Eixelberger, T.; Wolkenstein, G.; Hackner, R.; Bruns, V.; Mühldorfer, S.; Geissler, U.; Belle, S.; Wittenberg, T. YOLO networks for polyp detection: A human-in-the-loop training approach. Curr. Dir. Biomed. Eng. 2022, 8, 277–280. [Google Scholar] [CrossRef]
- Doniyorjon, M.; Madinakhon, R.; Shakhnoza, M.; Cho, Y.-I. An Improved Method of Polyp Detection Using Custom YOLOv4-Tiny. Appl. Sci. 2022, 12, 10856. [Google Scholar] [CrossRef]
- Zhang, R.; Zheng, Y.; Poon, C.C.; Shen, D.; Lau, J.Y. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recognit. 2018, 83, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.S.C.; Venkatesh, C.; Sinha, S.; Mazumdar, S. Real time Automatic Polyp Detection in White light Endoscopy videos using a combination of YOLO and DeepSORT. In Proceedings of the PCEMS 2022—1st International Conference on the Paradigm Shifts in Communication, Embedded Systems. Machine Learning and Signal Processing, Nagpur, India, 6–7 May 2022; pp. 104–106. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Z.; Song, L.; Liu, X.; Liu, S. Multiclassification of Endoscopic Colonoscopy Images Based on Deep Transfer Learning. Comput. Math Methods Med. 2021, 2021, 2485934. [Google Scholar] [CrossRef]
- NVenkatayogi, N.; Kara, O.C.; Bonyun, J.; Ikoma, N.; Alambeigi, F. Classification of Colorectal Cancer Polyps via Transfer Learning and Vision-Based Tactile Sensing. In Proceedings of the IEEE Sensors, Dallas, TX, USA, 30 October–2 November 2022. [Google Scholar] [CrossRef]
- Ribeiro, E.; Uhl, A.; Wimmer, G.; Häfner, M. Exploring Deep Learning and Transfer Learning for Colonic Polyp Classification. Comput. Math Methods Med. 2016, 2016, 6584725. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, D.; Zhang, C.; Zhang, P.; Xiong, Z.; Cao, Y.; Liu, B.; Liu, X.; Chen, S. Colorectal Polyp Detection in Real-world Scenario: Design and Experiment Study. In Proceedings of the International Conference on Tools with Artificial Intelligence, ICTAI, Baltimore, MD, USA, 9–11 November 2020; pp. 706–713. [Google Scholar] [CrossRef]
- Usami, H.; Iwahori, Y.; Adachi, Y.; Bhuyan, M.; Wang, A.; Inoue, S.; Ebi, M.; Ogasawara, N.; Kasugai, K. Colorectal Polyp Classification Based On Latent Sharing Features Domain from Multiple Endoscopy Images. Procedia Comput. Sci. 2020, 176, 2507–2514. [Google Scholar] [CrossRef]
- Bora, K.; Bhuyan, M.K.; Kasugai, K.; Mallik, S.; Zhao, Z. Computational learning of features for automated colonic polyp classification. Sci. Rep. 2021, 11, 4347. [Google Scholar] [CrossRef]
- Tashk, A.; Herp, J.; Nadimi, E.; Sahin, K.E. A CNN Architecture for Detection and Segmentation of Colorectal Polyps from CCE Images. In Proceedings of the 2022 IEEE 5th International Conference on Image Processing Applications and Systems (IPAS), Genova, Italy, 5–7 December 2022. Accepted/In press. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Gwak, J. Ensemble of Instance Segmentation Models for Polyp Segmentation in Colonoscopy Images. IEEE Access. 2019, 7, 26440–26447. [Google Scholar] [CrossRef]
- Liew, W.S.; Tang, T.B.; Lin, C.-H.; Lu, C.-K. Automatic colonic polyp detection using integration of modified deep residual convolutional neural network and ensemble learning approaches. Comput. Methods Programs Biomed. 2021, 206, 106114. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar] [CrossRef] [Green Version]
- Jinsakul, N.; Tsai, C.-F.; Tsai, C.-E.; Wu, P. Enhancement of Deep Learning in Image Classification Performance Using Xception with the Swish Activation Function for Colorectal Polyp Preliminary Screening. Mathematics 2019, 7, 1170. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Younas, F.; Usman, M.; Yan, W.Q. A deep ensemble learning method for colorectal polyp classification with optimized network parameters. Appl. Intell. 2022, 53, 2410–2433. [Google Scholar] [CrossRef]
- Liew, W.S.; Tang, T.B.; Lu, C.-K. Computer-aided diagnostic tool for classification of colonic polyp assessment. In Proceedings of the International Conference on Artificial Intelligence for Smart Community, Seri Iskandar, Malaysia, 17–18 December 2022; pp. 735–743. [Google Scholar] [CrossRef]
- Lo, C.-M.; Yeh, Y.-H.; Tang, J.-H.; Chang, C.-C.; Yeh, H.-J. Rapid Polyp Classification in Colonoscopy Using Textural and Convolutional Features. Healthcare 2022, 10, 1494. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Balabantaray, B.K.; Bora, K.; Mallik, S.; Kasugai, K.; Zhao, Z. An Ensemble-Based Deep Convolutional Neural Network for Computer-Aided Polyps Identification from Colonoscopy. Front. Genet. 2022, 13, 844391. [Google Scholar] [CrossRef]
- Rani, N.; Verma, R.; Jinda, A. Polyp Detection Using Deep Neural Networks. In Handbook of Intelligent Computing and Optimization for Sustainable Development; Scrivener Publishing LLC: Beverly, MA, USA, 2022; pp. 801–814. [Google Scholar] [CrossRef]
- Albuquerque, C.; Henriques, R.; Castelli, M. A stacking-based artificial intelligence framework for an effective detection and localization of colon polyps. Sci. Rep. 2022, 12, 17678. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Liao, H.-Y.M.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. CSPNet: A new backbone that can enhance learning capability of CNN. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 1571–1580. [Google Scholar] [CrossRef]
- Park, K.-B.; Lee, J.Y. SwinE-Net: Hybrid deep learning approach to novel polyp segmentation using convolutional neural network and Swin Transformer. J. Comput. Des. Eng. 2022, 9, 616–632. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. Available online: https://github. (accessed on 24 December 2022).
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, ICML, Long Beach, CA, USA, 9–15 June 2019; pp. 10691–10700. [Google Scholar] [CrossRef]
- Ho, C.; Zhao, Z.; Chen, X.F.; Sauer, J.; Saraf, S.A.; Jialdasani, R.; Taghipour, K.; Sathe, A.; Khor, L.-Y.; Lim, K.-H.; et al. A promising deep learning-assistive algorithm for histopathological screening of colorectal cancer. Sci. Rep. 2022, 12, 2222. [Google Scholar] [CrossRef]
- Wesp, P.; Grosu, S.; Graser, A.; Maurus, S.; Schulz, C.; Knösel, T.; Fabritius, M.P.; Schachtner, B.; Yeh, B.M.; Cyran, C.C.; et al. Deep learning in CT colonography: Differentiating premalignant from benign colorectal polyps. Eur. Radiol. 2022, 32, 4749–4759. [Google Scholar] [CrossRef] [PubMed]
- Biffi, C.; Salvagnini, P.; Dinh, N.N.; Hassan, C.; Sharma, P.; Antonelli, G.; Awadie, H.; Bernhofer, S.; Carballal, S.; Dinis-Ribeiro, M.; et al. A novel AI device for real-time optical characterization of colorectal polyps. NPJ Digit. Med. 2022, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- GI GeniusTM Intelligent Endoscopy Module | Medtronic. Available online: https://www.medtronic.com/covidien/en-us/products/gastrointestinal-artificial-intelligence/gi-genius-intelligent-endoscopy.html (accessed on 24 December 2022).
- Ellahyani, A.; El Jaafari, I.; Charfi, S.; El Ansari, M. Fine-tuned deep neural networks for polyp detection in colonoscopy images. Pers. Ubiquitous Comput. 2022. [Google Scholar] [CrossRef]
- Grosu, S.; Wesp, P.; Graser, A.; Maurus, S.; Schulz, C.; Knösel, T.; Cyran, C.C.; Ricke, J.; Ingrisch, M.; Kazmierczak, P.M. Machine learning-based differentiation of benign and premalignant colorectal polyps detected with CT colonography in an asymptomatic screening population: A proof-of-concept study. Radiology 2021, 299, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Jiang, H.; Ma, L.; Chang, Y. A Real-Time Polyp Detection Framework for Colonoscopy Video; Springer International Publishing: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Eu, C.Y.; Tang, T.B.; Lu, C.-K. Automatic Polyp Segmentation in Colonoscopy Images Using Single Network Model: SegNet; Springer Nature: Singapore, 2022. [Google Scholar] [CrossRef]
- Carrinho, P.; Falcao, G. Highly accurate and fast YOLOv4-Based polyp detection. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Yu, T.; Lin, N.; Zhang, X.; Pan, Y.; Hu, H.; Zheng, W.; Liu, J.; Hu, W.; Duan, H.; Si, J. An end-to-end tracking method for polyp detectors in colonoscopy videos. Artif. Intell. Med. 2022, 131, 102363. [Google Scholar] [CrossRef]
| Types | Average Size | Cancerous | Frequency in Patients |
|---|---|---|---|
| Tubular adenomas | 1 to 12 mm | Non-cancerous | 50% |
| Villous adenomas | ≥20 mm | Cancerous | 5% |
| Tubulovillous adenomas | ≤1 cm | Cancerous | 8–16% |
| Serrated adenomas | ≤10 mm | Non-cancerous | 15–20% |
| Hyperplastic | ≤5 mm | Non-cancerous | 20–40% |
| Inflammatory | ≤2 cm | Non-cancerous | 10–20% |
| Name | Number of Samples | Resolution | Media Type |
|---|---|---|---|
| Kvasir-SEG | 1000 | Between 332 × 487 and 1920 × 1072 | Static images |
| ETIS-Larib | 196 | 1225 × 996 | Static images |
| CVC-ClinicDB | 612 | 384 × 288 | Static images |
| CVC-PolypHD | 56 | 1920 × 1080 | Static images |
| CVC-ColonDB | 300 | 574 × 500 | Static images |
| EndoTect | 110,709 images + 373 videos | - | Static images/videos |
| BKAI-IGH NeoPolyp-Small | 1200 | - | Static images |
| NeoPolyp | 7500 | - | Static images |
| PolypGen | 8037 | - | Static images |
| EndoScene | 912 | 384 × 288 (CVC-ClinicDB) 574 × 500 (CVC-ColonDB) | Static images |
| SUN | 49,136 | - | Video |
| EndoTest | 48 | - | Video |
| ASU-Mayo clinic colonoscopy | 38 | - | Video |
| Colonoscopic Dataset | 76 | 768 × 576 | Video |
| CVC-ClinicVideoDB | 18 | 768 × 576 | Video |
| Method | Precision | Recall | F1 | IoU | Accuracy | Sens. | Spec | AUC | Training Set | Testing Set |
|---|---|---|---|---|---|---|---|---|---|---|
| [8] | 90.61 | 91.04 | 90.82 | - | - | - | - | - | SUN, PICCOLO Widefield | ETIS -Larib |
| [13] | 92.30 | - | 92.40 | - | - | - | - | - | EndoScene, Kvasir-SEG | EndoScene, Kvasir-SEG |
| [10] | 94.9 | 96.9 | 95.9 | - | - | - | - | - | CVC-ClinicDB, ETIS-Larib, Kvasir-SEG | Private dataset |
| [9] | 100 | 99.20 | 99.60 | - | - | - | - | - | CVC-ClincDB | CVC-ColonDB, ETIS-Larib |
| [14] | 87.4 | 84.4 | 85.9 | - | - | - | - | - | ASU-Mayo | ASU-Mayo |
| [58] | - | - | - | - | 92% | - | - | - | University of Leeds dataset | University of Leeds dataset |
| [82] | 98.6 | 98.01 | - | - | - | - | - | - | Private dataset, Kvasir-SEG | Private dataset, Kvasir-SEG |
| [11] | 78.5 | 78.8 | 78.6 | - | - | - | - | - | Private dataset | Private dataset |
| [86] | - | - | 88.0 | 84.2 | - | - | - | - | Kvasir-SEG, CVC-ClinicDB | CVC-ColonDB, ETIS-Larib, EndoScene |
| [89] | - | - | 97.4 | - | - | 97.4 | 60.3 | 91.7 | Private dataset | Private dataset |
| [90] | - | - | - | - | - | - | - | 83.0 | Cancer Imaging Archive | Cancer Imaging Archive |
| [91] | - | - | - | - | 84.8% | 80.7 | 87.3 | - | Private dataset | Private dataset |
| [93] | 91.9 | 89.0 | 90.0 | - | - | - | - | - | Kvasir-SEG | CVC-ClinicDB, ETIS-Larib |
| [16] | 89.0 | 87.0 | 88.0 | - | - | - | - | - | Private dataset | Private dataset |
| [94] | - | - | - | - | - | 82.0 | 85.0 | 91.0 | Private dataset | Private dataset |
| [95] | 83.6 | 73.1 | 78.0 | - | - | - | - | - | CVC-ClinicDB, ETIS-Larib, CVC-ClinicVideoDB | CVC-Clinic, ETIS-Larib, CVC-ClinicVideo |
| [96] | - | - | - | 81.7 | - | - | - | - | CVC-ClinicDB, CVC-ColonDB, ETIS-Larib | CVC-Clinic, CVC-ColonDB, ETIS-Larib |
| [97] | 80.53 | 73.56 | 76.88 | - | - | - | - | - | Kvasir-SEG, CVC-ClinicDB, CVC-ColonDB | Kvasir-SEG, CVC-Clinic, CVC-ColonDB |
| [98] | 92.60 | 80.70 | 86.24 | - | - | - | - | - | CVC-ClinicDB, CVC-VideoClinicDB | ETIS-Larib |
| [12] | 93.45 | - | 89.65 | - | - | 86.14 | 85.32 | - | SUN, Kvasir-SEG | SUN, Kvasir-SEG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
ELKarazle, K.; Raman, V.; Then, P.; Chua, C. Detection of Colorectal Polyps from Colonoscopy Using Machine Learning: A Survey on Modern Techniques. Sensors 2023, 23, 1225. https://doi.org/10.3390/s23031225
ELKarazle K, Raman V, Then P, Chua C. Detection of Colorectal Polyps from Colonoscopy Using Machine Learning: A Survey on Modern Techniques. Sensors. 2023; 23(3):1225. https://doi.org/10.3390/s23031225
Chicago/Turabian StyleELKarazle, Khaled, Valliappan Raman, Patrick Then, and Caslon Chua. 2023. "Detection of Colorectal Polyps from Colonoscopy Using Machine Learning: A Survey on Modern Techniques" Sensors 23, no. 3: 1225. https://doi.org/10.3390/s23031225
APA StyleELKarazle, K., Raman, V., Then, P., & Chua, C. (2023). Detection of Colorectal Polyps from Colonoscopy Using Machine Learning: A Survey on Modern Techniques. Sensors, 23(3), 1225. https://doi.org/10.3390/s23031225






