A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria
Abstract
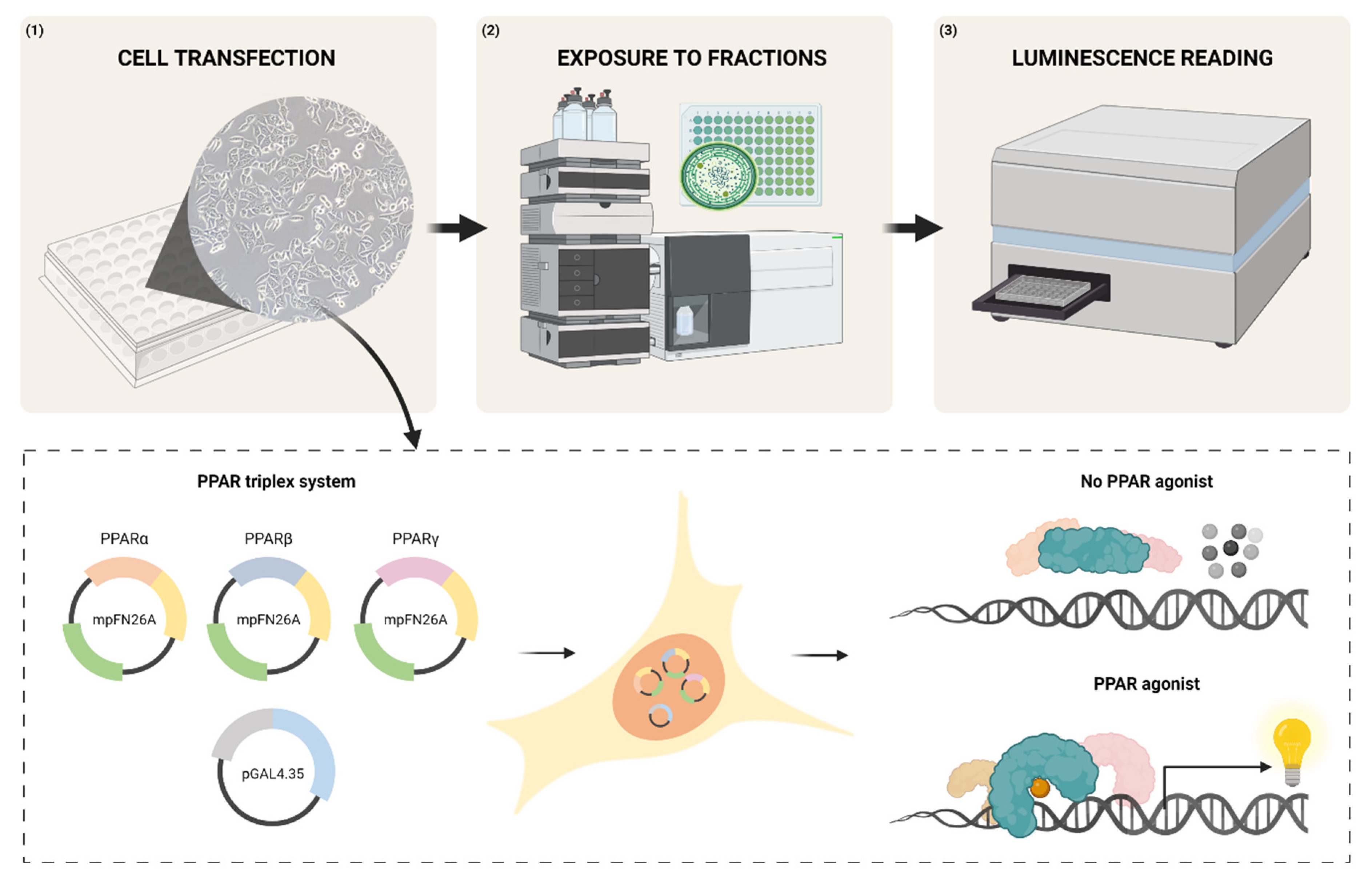
:1. Introduction
2. Materials and Methods
2.1. Construction of Plasmid Vectors
2.2. Isolation and Cloning of PPARα, -β and -γ
2.3. Transactivation Assays
2.3.1. Optimization of Vectors Ratio in mpFN26A[Fluc]/mpGL4.35[Nluc] System
2.3.2. Characterization of mpFN26A[Fluc]/mpGL4.35[Nluc] Sensor System
2.3.3. Response of pBIND[Rluc]/pGL4.35[Fluc] Sensor System to PPARs’ Reference Agonists
2.3.4. Comparison of pBIND[Rluc]/pGL4.35[Fluc] and mpFN26A[Fluc]/mpGL4.35[Nluc] Systems
2.3.5. Specificity of mpFN26A[Fluc]/mpGL4.35[Nluc] System
2.4. Culture of Cyanobacteria and Production of Crude Extracts and Fractions
Screening of PPARs’ Ligands in Cyanobacteria Fractions
2.5. Data Analysis and Statistics
3. Results and Discussion
3.1. Optimization of Vector Ratio in mpFN26A[Fluc]/mpGL4.35[Nluc] System
3.2. Characterization of mpFN26A[Fluc]/mpGL4.35[Nluc] Sensor System
3.3. Comparison of pBIND[Rluc]/pGL4.35[Fluc] and mpFN26A[Fluc]/mpGL4.35[Nluc] Systems
3.4. Specificity of mpFN26A[Fluc]/mpGL4.35[Nluc] System
3.5. Screening of PPARs Ligands in Cyanobacteria Fractions
3.6. Low Firefly Luciferase (Fluc) Raw Values Observed with Some Fractions Assayed
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Auwerx, J.; Baulieu, E.; Beato, M.; Becker-Andre, M.; Burbach, P.H.; Camerino, G.; Chambon, P.; Cooney, A.; Dejean, A.; Dreyer, C.; et al. A Unified Nomenclature System for the Nuclear Receptor Superfamily. Cell 1999, 97, 161–163. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Gupta, P.; Saini, A.; Kaushal, C.; Sharma, S. The Peroxisome Proliferator-Activated Receptor: A Family of Nuclear Receptors Role in Various Diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARs in Health and Disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Reddy, J.K. Transcription Coactivators for Peroxisome Proliferator-Activated Receptors. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2007, 1771, 936–951. [Google Scholar] [CrossRef] [PubMed]
- Aranda, A.; Pascual, A. Nuclear Hormone Receptors and Gene Expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [CrossRef] [Green Version]
- Zoete, V.; Grosdidier, A.; Michielin, O. Peroxisome Proliferator-Activated Receptor Structures: Ligand Specificity, Molecular Switch and Interactions with Regulators. Biochim. Biophys. Acta 2007, 1771, 915–925. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Forman, B.M.; Blumberg, B.; Ong, E.S.; Borgmeyer, U.; Mangelsdorf, D.J.; Umesono, K.; Evans, R.M. Differential Expression and Activation of a Family of Murine Peroxisome Proliferator-Activated Receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 7355–7359. [Google Scholar] [CrossRef] [Green Version]
- Wagner, N.; Wagner, K.D. The Role of PPARs in Disease. Cells 2020, 9, 2367. [Google Scholar] [CrossRef]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR Control of Metabolism and Cardiovascular Functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef]
- Tysoe, O. PPAR Mediates Intestinal Stem Cell Tumorigenesis. Nat. Rev. Endocrinol. 2021, 17, 514. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, M.; Codocedo, J.F.; Puntambekar, S.S.; Landreth, G.E. Nuclear Receptors as Therapeutic Targets for Neurodegenerative Diseases: Lost in Translation. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Wójtowicz, S.; Strosznajder, A.K.; Jeżyna, M.; Strosznajder, J.B. The Novel Role of PPAR Alpha in the Brain: Promising Target in Therapy of Alzheimer’s Disease and Other Neurodegenerative Disorders. Neurochem. Res. 2020, 45, 972–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azhar, S. Peroxisome Proliferator-Activated Receptors, Metabolic Syndrome and Cardiovascular Disease. Future Cardiol. 2010, 6, 657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmon, G.S.; Lam, M.T.; Glass, C.K. PPARs and Lipid Ligands in Inflammation and Metabolism. Chem. Rev. 2011, 111, 6321. [Google Scholar] [CrossRef] [Green Version]
- Jones, D. Potential Remains for PPAR-Targeted Drugs. Nat. Rev. Drug Discov. 2010, 9, 668–669. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARα. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef]
- Edin, M.L.; Lih, F.B.; Hammock, B.D.; Thomson, S.; Zeldin, D.C.; Bishop-Bailey, D. Vascular Lipidomic Profiling of Potential Endogenous Fatty Acid PPAR Ligands Reveals the Coronary Artery as Major Producer of CYP450-Derived Epoxy Fatty Acids. Cells 2020, 9, 1096. [Google Scholar] [CrossRef]
- Bishop-Bailey, D.; Wray, J. Peroxisome Proliferator-Activated Receptors: A Critical Review on Endogenous Pathways for Ligand Generation. Prostaglandins Other Lipid Mediat. 2003, 71, 1–22. [Google Scholar] [CrossRef]
- Singh, R.; Parihar, P.; Singh, M.; Bajguz, A.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Uncovering Potential Applications of Cyanobacteria and Algal Metabolites in Biology, Agriculture and Medicine: Current Status and Future Prospects. Front. Microbiol. 2017, 8, 515. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, A.K.; Tiwari, B.S. Cyanotherapeutics: An Emerging Field for Future Drug Discovery. Appl. Phycol. 2020, 1, 44–57. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Kruse, O.; Hellingwerf, K.J. Potential of Industrial Biotechnology with Cyanobacteria and Eukaryotic Microalgae. Curr. Opin. Biotechnol. 2013, 24, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalifa, S.A.M.; Shedid, E.S.; Saied, E.M.; Jassbi, A.R.; Jamebozorgi, F.H.; Rateb, M.E.; Du, M.; Abdel-Daim, M.M.; Kai, G.Y.; Al-Hammady, M.A.M.; et al. Cyanobacteria—From the Oceans to the Potential Biotechnological and Biomedical Applications. Mar. Drugs 2021, 19, 241. [Google Scholar] [CrossRef]
- Ramos, V.; Morais, J.; Castelo-Branco, R.; Pinheiro, Â.; Martins, J.; Regueiras, A.; Pereira, A.L.; Lopes, V.R.; Frazão, B.; Gomes, D.; et al. Cyanobacterial Diversity Held in Microbial Biological Resource Centers as a Biotechnological Asset: The Case Study of the Newly Established LEGE Culture Collection. J. Appl. Phycol. 2018, 30, 1437–1451. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, S.A.C.; Preto, M.; Moreira, G.; Martins, T.P.; Abt, K.; Melo, A.; Vasconcelos, V.M.; Leão, P.N. Discovery of Cyanobacterial Natural Products Containing Fatty Acid Residues**. Angew. Chem. Int. Ed. 2021, 60, 10064–10072. [Google Scholar] [CrossRef] [PubMed]
- Tidgewell, K.; Clark, B.R.; Gerwick, W.H. The Natural Products Chemistry of Cyanobacteria. Compr. Nat. Prod. II Chem. Biol. 2010, 2, 141–188. [Google Scholar] [CrossRef]
- Martins, T.P.; Rouger, C.; Glasser, N.R.; Freitas, S.; De Fraissinette, N.B.; Balskus, E.P.; Tasdemir, D.; Leão, P.N. Chemistry, Bioactivity and Biosynthesis of Cyanobacterial Alkylresorcinols. Nat. Prod. Rep. 2019, 36, 1437–1461. [Google Scholar] [CrossRef] [PubMed]
- Solomon, C.; White, J.H.; Kremer, R. Mitogen-Activated Protein Kinase Inhibits 1,25-Dihydroxyvitamin D3–Dependent Signal Transduction by Phosphorylating Human Retinoid X Receptor α. J. Clin. Investig. 1999, 103, 1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muralikumar, S.; Vetrivel, U.; Narayanasamy, A.; Das, U.N. Probing the Intermolecular Interactions of PPARγ-LBD with Polyunsaturated Fatty Acids and Their Anti-Inflammatory Metabolites to Infer Most Potential Binding Moieties. Lipids Health Dis. 2017, 16, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruzeiro, C.; Lopes-Marques, M.; Ruivo, R.; Rodrigues-Oliveira, N.; Santos, M.M.; Rocha, M.J.; Rocha, E.; Castro, L.F.C. A Mollusk VDR/PXR/CAR-like (NR1J) Nuclear Receptor Provides Insight into Ancient Detoxification Mechanisms. Aquat. Toxicol. 2016, 174, 61–69. [Google Scholar] [CrossRef]
- Capitão, A.M.F.; Lopes-Marques, M.S.; Ishii, Y.; Ruivo, R.; Fonseca, E.S.S.; Páscoa, I.; Jorge, R.P.; Barbosa, M.A.G.; Hiromori, Y.; Miyagi, T.; et al. Evolutionary Exploitation of Vertebrate Peroxisome Proliferator-Activated Receptor γ by Organotins. Environ. Sci. Technol. 2018, 52, 13951–13959. [Google Scholar] [CrossRef] [PubMed]
- Videira, N.B.; Batista, F.A.H.; Torres Cordeiro, A.; Figueira, A.C.M. Cellular and Biophysical Pipeline for the Screening of Peroxisome Proliferator-Activated Receptor Beta/Delta Agonists: Avoiding False Positives. PPAR Res. 2018, 2018, 3681590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filho, H.V.R.; Videira, N.B.; Bridi, A.V.; Tittanegro, T.H.; Batista, F.A.H.; de Carvalho Pereira, J.G.; de Oliveira, P.S.L.; Bajgelman, M.C.; Le Maire, A.; Figueira, A.C.M. Screening for PPAR Non-Agonist Ligands Followed by Characterization of a Hit, AM-879, with Additional No-Adipogenic and Cdk5-Mediated Phosphorylation Inhibition Properties. Front. Endocrinol. Lausanne 2018, 9, 1. [Google Scholar] [CrossRef]
- Bai, C.; Schmidt, A.; Freedman, L.P. Steroid Hormone Receptors and Drug Discovery: Therapeutic Opportunities and Assay Designs. Assay Drug Dev. Technol. 2003, 1, 843–852. [Google Scholar] [CrossRef]
- Paguio, A.; Stecha, P.; Wood, K.V.; Fan, F. Improved Dual-Luciferase Reporter Assays for Nuclear Receptors. Curr. Chem. Genomics 2010, 4, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auld, D.S.; Narahari, J.; Ho, P.I.; Casalena, D.; Nguyen, V.; Cirbaite, E.; Hughes, D.; Daly, J.; Webb, B. Characterization and Use of TurboLuc Luciferase as a Reporter for High-Throughput Assays. Biochemistry 2018, 57, 4700–4706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, M.; Ozawa, T. Advanced Bioluminescence System for In Vivo Imaging with Brighter and Red-Shifted Light Emission. Int. J. Mol. Sci. 2020, 21, 6538. [Google Scholar] [CrossRef]
- Grover, G.S.; Turner, B.A.; Parker, C.N.; Meier, J.; Lala, D.S.; Lee, P.H. Multiplexing Nuclear Receptors for Agonist Identification in a Cell-Based Reporter Gene High-Throughput Screen. J. Biomol. Screen 2003, 8, 239–246. [Google Scholar] [CrossRef] [Green Version]
- André, A.; Ruivo, R.; Capitão, A.; Froufe, E.; Páscoa, I.; Costa Castro, L.F.; Santos, M.M. Cloning and Functional Characterization of a Retinoid X Receptor Orthologue in Platynereis Dumerilii: An Evolutionary and Toxicological Perspective. Chemosphere 2017, 182, 753–761. [Google Scholar] [CrossRef]
- Páscoa, I.; Fonseca, E.; Ferraz, R.; Machado, A.M.; Conrado, F.; Ruivo, R.; Cunha, I.; Castro, L.F.C. The Preservation of PPARg; Genome Duplicates in Some Teleost Lineages: Insights into Lipid Metabolism and Xenobiotic Exploitation. Genes 2022, 13, 107. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Morais, J.; Preto, M.; Silva, R.; Urbatzka, R.; Vasconcelos, V.; Reis, M. Uncovering the Bioactive Potential of a Cyanobacterial Natural Products Library Aided by Untargeted Metabolomics. Mar. Drugs 2021, 19, 633. [Google Scholar] [CrossRef] [PubMed]
- Pavek, P. Pregnane X Receptor (PXR)-Mediated Gene Repression and Cross-Talk of PXR with Other Nuclear Receptors via Coactivator Interactions. Front. Pharmacol. 2016, 7, 456. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Mella-Herrera, R.A.; Golden, J.W. Cyanobacterial Heterocysts. Cold Spring Harb. Perspect. Biol. 2010, 2, a000315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzl, T.; Haedrich, J.; Schaechtele, A.; Robouch, P.; Stroka, J. JRC Publications Repository-Guidance Document on the Estimation of LOD and LOQ for Measurements in the Field of Contaminants in Feed and Food; Publications Office of the European Union: Luxembourg, 2016; Volume JRC102946. [Google Scholar]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29, S49. [Google Scholar]
- ICH Topic Q 2 (R1) Validation of Analytical Procedures: Text and Methodology Step 5-Note for Guidance on Validation of Analytical Procedures: Text and Methodology (CPMP/ICH/381/95). In Proceedings of the International Conference on Harmonization; European Medicin Agenci (EMEA), Ed.; 1995; pp. 1–15. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-2-r1-validation-analytical-procedures-text-methodology-step-5_en.pdf (accessed on 7 December 2022).
- Guidelines for the Validation and Verification of Quantitative and Qualitative Test Methods; National Association of Testing Authorities, Australia. 2012. Available online: http://www.demarcheiso17025.com/document/Guidelines%20for%20the%20validation%20and%20verification%20of%20quantitative%20and%20qualitative%20test%20methods.pdf (accessed on 7 December 2022).
- Sebaugh, J.L. Guidelines for Accurate EC50/IC50 Estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Di Veroli, G.Y.; Fornari, C.; Goldlust, I.; Mills, G.; Koh, S.B.; Bramhall, J.L.; Richards, F.M.; Jodrell, D.I. An Automated Fitting Procedure and Software for Dose-Response Curves with Multiphasic Features. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Goktug, A.N.; Chai, S.C.; Chen, T. Data Analysis Approaches in High Throughput Screening. In Drug Discovery; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.H.; Chung, T.D.Y.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Garikapati, K.K.; Ravi, R.K.; Krishnamurthy, P.T.; Narenderan, S.T.; Babu, B.; Nagappan, K. Quantification of Rosiglitazone in Rat Plasma and Tissues via LC-MS/MS: Method Development, Validation, and Application in Pharmacokinetic and Tissue Distribution Studies. Biomed. Chromatogr. 2022, 36, e5326. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, A.; Moeser, M.; Medvedeva, L.; Martsen, E.; Granick, A.; Raines, L.; Gorman, K.; Lin, B.; Zeng, M.; Houck, K.A.; et al. Comprehensive Assessment of NR Ligand Polypharmacology by a Multiplex Reporter NR Assay. Sci. Rep. 2022, 12, 3115. [Google Scholar] [CrossRef]
- Seimandi, M.; Lemaire, G.; Pillon, A.; Perrin, A.; Carlavan, I.; Voegel, J.J.; Vignon, F.; Nicolas, J.C.; Balaguer, P. Differential Responses of PPARalpha, PPARdelta, and PPARgamma Reporter Cell Lines to Selective PPAR Synthetic Ligands. Anal. Biochem. 2005, 344, 8–15. [Google Scholar] [CrossRef]
- Lopes, G.; Clarinha, D.; Vasconcelos, V. Carotenoids from Cyanobacteria: A Biotechnological Approach for the Topical Treatment of Psoriasis. Microorganisms 2020, 8, 302. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Rosa, F.; Ribeiro, T.; Hernandez-Bautista, R.; Bonaldo, M.; Silva, N.G.; Eiríksson, F.; Thorsteinsdóttir, M.; Ussar, S.; Urbatzka, R. Identification of Cyanobacterial Strains with Potential for the Treatment of Obesity-Related Co-Morbidities by Bioactivity, Toxicity Evaluation and Metabolite Profiling. Mar. Drugs 2019, 17, 280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, V.R.; Schmidtke, M.; Fernandes, M.H.; Martins, R.; Vasconcelos, V. Cytotoxicity in L929 Fibroblasts and Inhibition of Herpes Simplex Virus Type 1 Kupka by Estuarine Cyanobacteria Extracts. Toxicol. Vitr. 2011, 25, 944–950. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Garcia, M.; Costa-Rodrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring Bioactive Properties of Marine Cyanobacteria Isolated from the Portuguese Coast: High Potential as a Source of Anticancer Compounds. Mar. Drugs 2014, 12, 98–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giulia, C.; Adriana, L.; Elisabetta, P.; Poli, G.; Elisabetta, C.; Sara, M.; Tonino, E.; Andrea, G.; Mario, S.; Massimo, M.; et al. Rosiglitazone Inhibits Adrenocortical Cancer Cell Proliferation by Interfering with the IGF-IR Intracellular Signaling. PPAR Res. 2008, 2008, 904041. [Google Scholar] [CrossRef] [Green Version]
- Rondón-Ortiz, A.N.; Cardenas, C.L.L.; Martínez-Málaga, J.; Gonzales-Urday, A.L.; Gugnani, K.S.; Böhlke, M.; Maher, T.J.; Pino-Figueroa, A.J. High Concentrations of Rosiglitazone Reduce MRNA and Protein Levels of LRP1 in HepG2 Cells. Front. Pharmacol. 2017, 8, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, T.; Lemos, F.; Preto, M.; Azevedo, J.; Sousa, M.L.; Leão, P.N.; Campos, A.; Linder, S.; Vitorino, R.; Vasconcelos, V.; et al. Cytotoxicity of Portoamides in Human Cancer Cells and Analysis of the Molecular Mechanisms of Action. PLoS ONE 2017, 12, e0188817. [Google Scholar] [CrossRef] [Green Version]
- Kenda, M.; Vegelj, J.; Herlah, B.; Perdih, A.; Mladěnka, P.; Sollner Dolenc, M. Evaluation of Firefly and Renilla Luciferase Inhibition in Reporter-Gene Assays: A Case of Isoflavonoids. Int. J. Mol. Sci. 2021, 22, 6927. [Google Scholar] [CrossRef]





| Compound | GW7647 (PPARα Agonist) | GW501516 (PPARβ Agonist) | Rosiglitazone (PPARγ Agonist) | |||||
|---|---|---|---|---|---|---|---|---|
| Mode | Uniplex | Triplex | Uniplex | Triplex | Uniplex | Triplex | Uniplex pBIND/pGL4.35 | |
| EC50 (nM) | 12.97 | - | 3.59 | - | 29.37 | - | - | |
| Linear equation Parameters * | a | 7.565 | −3.084 | 20.31 | 1.541 | −35.43 | −30.94 | −3.084 |
| b | 28.71 | 24.86 | 41.20 | 24.59 | 54.83 | 32.87 | 24.86 | |
| R2 | 0.948 | 0.981 | 0.929 | 0.933 | 0.941 | 0.912 | 0.980 | |
| SEy response (nM) | 8.631 | 5.657 | 9.416 | 8.482 | 5.017 | 11.04 | 5.656 | |
| LOD (nM) | 1.173 | 0.887 | 0.891 | 1.345 | 0.357 | 1.310 | 0.887 | |
| LOQ (nM) | 3.870 | 2.928 | 2.942 | 4.439 | 1.178 | 4.322 | 2.928 | |
| LRD (nM) | [1;1000] | [1;10,000] | [0.4;100] | [1;10,000] | [6.4;100] | [1.6;10,000] | [10;1000] | |
| Uniplex | Triplex | |||
|---|---|---|---|---|
| PPARα | PPARβ | PPARγ | PPARα, -β and -γ | |
| DMSO | 0.987 ± 0.025 | 1.026 ± 0.030 | 0.975 ± 0.040 | 1.042 ± 0.010 |
| Clotrimazole | 0.944 ± 0.051 | 1.167 ± 0.085 | 0.976 ± 0.114 | 1.15 ± 0.35 |
| p-value | 0.871 | 0.156 | 0.239 | 0.574 |
| Order | Strain | Taxon | Fraction | FI Confirmation |
|---|---|---|---|---|
| Nostocales | LEGE 02266 | Sphaerospermopsis sp. | D | 2.448 |
| E | 2.420 | |||
| Oscillatoriales | LEGE 06078 | Oxynema acuminatum | F | 3.003 |
| LEGE 06139 | Cyanobium sp. | B | 2.229 | |
| LEGE 06141 | Oculatella sp. | G | 0.494 | |
| LEGE 13457 | Nodosilinea antarctica | A | 0.489 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Páscoa, I.; Biltes, R.; Sousa, J.; Preto, M.A.C.; Vasconcelos, V.; Castro, L.F.; Ruivo, R.; Cunha, I. A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria. Sensors 2023, 23, 1338. https://doi.org/10.3390/s23031338
Páscoa I, Biltes R, Sousa J, Preto MAC, Vasconcelos V, Castro LF, Ruivo R, Cunha I. A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria. Sensors. 2023; 23(3):1338. https://doi.org/10.3390/s23031338
Chicago/Turabian StylePáscoa, Inês, Rita Biltes, João Sousa, Marco Aurélio Correia Preto, Vitor Vasconcelos, Luís Filipe Castro, Raquel Ruivo, and Isabel Cunha. 2023. "A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria" Sensors 23, no. 3: 1338. https://doi.org/10.3390/s23031338
APA StylePáscoa, I., Biltes, R., Sousa, J., Preto, M. A. C., Vasconcelos, V., Castro, L. F., Ruivo, R., & Cunha, I. (2023). A Multiplex Molecular Cell-Based Sensor to Detect Ligands of PPARs: An Optimized Tool for Drug Discovery in Cyanobacteria. Sensors, 23(3), 1338. https://doi.org/10.3390/s23031338









