Computer-Aided Ankle Ligament Injury Diagnosis from Magnetic Resonance Images Using Machine Learning Techniques
Abstract
:1. Introduction
- Stratify the possibilities of morphological variations on the ligament and its correction with ankle instability;
- Compare the ability for diagnosis by the magnetic resonance of different evaluators;
- To develop a method for extracting and classifying ankle ligaments to aid medical management;
- To compare and analyze different feature extraction techniques;
- Validate the results through statistical evaluations;
- To compare and analyze human diagnostic capability with software-based capability.
2. Materials and Methods
2.1. Ethical Statements
2.2. Patient Selection
2.3. Computational Characterization of the ATFL
2.4. Description of the Database
2.5. Data Extraction
2.5.1. Gray Level Co-Occurrence Matrix (GLCM)
2.5.2. Local Binary Patterns (LBP)
2.5.3. Hounsfield Unit Invariant Moments
2.5.4. Dimensional Characteristics (DC)
2.6. Classification Methods
2.6.1. Multi Layer Perceptron (MLP)
2.6.2. Support Vector Machine (SVM)
2.6.3. Random Forest (RF)
2.6.4. k-Nearest Neighbors (k-NN)
3. Experimental Setup and Performance Metrics
3.1. Validation Metrics
- True Positive (TP): The TP occurs when considering the real dataset, where the ATFL class was predicted correctly as the ATFL class;
- True Negative (TN): The TN occurs when considering the actual dataset, where the healthy control class was correctly predicted as the healthy control class;
- False Negative (FN): The FN occurs when considering the real set of data, where the class that is sought to be predicted was incorrectly predicted. This happens, when it was supposed to be ATFL and was classified as a healthy control;
- False Positive (FP): The FP occurs considering the real set of data, where the class that is sought to be predicted was incorrectly predicted. This happens, when it was supposed to be healthy control and was classified as ATFL.
- Accuracy: Refers to the global hit probability, which is the measure of general hit rate considering the two analyzed classes, considering errors and hits.
- F1-score: Refers to the harmonic mean between accuracy and recall. It is often used to evaluate unbalanced bases.
- ATFL class hit rate (ATFL): Refers to the probability that a patient who has a positive diagnosis for ATFL actually has ATFL.
- Healthy control class hit rate (HealthyControl): Refers to the probability of a patient who has a negative diagnosis for ATFL, that is, a patient from the healthy control class and that does not have ATFL.
3.2. Medical Analysis
4. Results and Discussion
4.1. Human Analysis
4.2. Computational Analysis
- Using the GLCM features improves the ACC Global measure when using the MLP classifier ();
- Using the HU features improves the ACC Global measure when using the RF classifier (), and the HealthyControl measure when using the MLP classifier ();
- SVM is never able to outperform significantly () other classifiers;
4.3. Comparative Evaluation between Human Analysis and Computational Analysis
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Conlin, F.D.; Johnson, P.G.; Sinning, J.E., Jr. The etiology and repair of rotary ankle instability. Foot Ankle 1989, 10, 152–155. [Google Scholar] [CrossRef]
- Bajuri, M.Y.; Daun, E.; Raof, M.H.A.; Hassan, M.R.; Das, S. Functional outcome of modified Brostrom-Gould procedure using the PopLok knotless suture anchor technique in lateral ankle instability. Cureus 2019, 11, e4971. [Google Scholar] [CrossRef]
- Milner, C.E.; Soames, R.W. Anatomy of the collateral ligaments of the human ankle joint. Foot Ankle Int. 1998, 19, 757–760. [Google Scholar] [CrossRef]
- Renström, A.F.H. and Scott A. Lynch Ankle ligament injuries. Rev. Bras. Med. Esporte 1998, 4, 71–80. [Google Scholar] [CrossRef]
- Tao, H.; Hu, Y.; Qiao, Y.; Ma, K.; Yan, X.; Hua, Y.; Chen, S. T2-mapping evaluation of early cartilage alteration of talus for chronic lateral ankle instability with isolated anterior talofibular ligament tear or combined with calcaneofibular ligament tear. J. Magn. Reson. Imaging 2018, 47, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Nagatomo, M.; Yoshimura, I.; Hagio, T.; Ishimatsu, T.; Sugino, Y.; Yamamoto, T. Straight Form of Calcaneofibular Ligament as a Three-Dimensional Magnetic Resonance Imaging Sign in Diagnosis of Calcaneofibular Ligament and Anterior Talofibular Ligament Inferior Fascicle Injury. J. Foot Ankle Surg. 2022, 61, 327–332. [Google Scholar] [CrossRef]
- Hall, E.A.; Docherty, C.L.; Simon, J.; Kingma, J.J.; Klossner, J.C. Strength-training protocols to improve deficits in participants with chronic ankle instability: A randomized controlled trial. J. Athl. Train. 2015, 50, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Sofka, C.M. Imaging Techniques for Assessment of Dynamically Unstable Sports Related Foot and Ankle Injuries. Foot Ankle Clin. 2021, 26, 205–224. [Google Scholar] [CrossRef]
- Noto, A.M.; Cheung, Y.; Rosenberg, Z.S.; Norman, A.; Leeds, N. MR imaging of the ankle: Normal variants. Radiology 1989, 170, 121–124. [Google Scholar] [CrossRef]
- Mengiardi, B.; Zanetti, M.; Schöttle, P.B.; Vienne, P.; Bode, B.; Hodler, J.; Pfirrmann, C.W. Spring ligament complex: MR imaging–anatomic correlation and findings in asymptomatic subjects. Radiology 2005, 237, 242–249. [Google Scholar] [CrossRef]
- Kim, H.K.; Laor, T.; Shire, N.J.; Bean, J.A.; Dardzinski, B.J. Anterior and posterior cruciate ligaments at different patient ages: MR imaging findings. Radiology 2008, 247, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.J.; Bohne, W.H.; Kaye, J.J. A radiographic study of the ligamentous anatomy of the ankle. Radiology 1977, 125, 659–667. [Google Scholar] [CrossRef]
- Elkaïm, M.; Thès, A.; Lopes, R.; Andrieu, M.; Cordier, G.; Molinier, F.; Benoist, J.; Colin, F.; Boniface, O.; Guillo, S.; et al. Agreement between arthroscopic and imaging study findings in chronic anterior talo-fibular ligament injuries. Orthop. Traumatol. Surg. Res. 2018, 104, S213–S218. [Google Scholar] [CrossRef] [PubMed]
- Barini, M.; Zagaria, D.; Licandro, D.; Pansini, S.; Airoldi, C.; Leigheb, M.; Carriero, A. Magnetic Resonance Accuracy in the Diagnosis of Anterior Talo-Fibular Ligament Acute Injury: A Systematic Review and Meta-Analysis. Diagnostics 2021, 11, 1782. [Google Scholar] [CrossRef]
- Liu, W.; Li, H.; Hua, Y. Quantitative magnetic resonance imaging (MRI) analysis of anterior talofibular ligament in lateral chronic ankle instability ankles pre-and postoperatively. BMC Musculoskelet. Disord. 2017, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, Y.B.; Kim, T.G.; Lee, S.W.; Park, S.H.; Lee, H.J.; Choi, Y.J.; Koh, Y.G. Reliability and validity of magnetic resonance imaging for the evaluation of the anterior talofibular ligament in patients undergoing ankle arthroscopy. Arthrosc. J. Arthrosc. Relat. Surg. 2015, 31, 1540–1547. [Google Scholar] [CrossRef]
- Cao, S.; Wang, C.; Ma, X.; Wang, X.; Huang, J.; Zhang, C. Imaging diagnosis for chronic lateral ankle ligament injury: A systemic review with meta-analysis. J. Orthop. Surg. Res. 2018, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Choi, J.G.; Jeong, B.O. The signal intensity of preoperative magnetic resonance imaging has predictive value for determining the arthroscopic reparability of the anterior talofibular ligament. Knee Surg. Sport. Traumatol. Arthrosc. 2021, 29, 1535–1543. [Google Scholar] [CrossRef]
- Gimber, L.H.; Latt, L.D.; Caruso, C.; Zuniga, A.A.N.; Krupinski, E.A.; Klauser, A.S.; Taljanovic, M.S. Ultrasound shear wave elastography of the anterior talofibular and calcaneofibular ligaments in healthy subjects. J. Ultrason. 2021, 21, 86–94. [Google Scholar] [CrossRef]
- Cordier, G.; Nunes, G.A.; Vega, J.; Roure, F.; Dalmau-Pastor, M. Connecting fibers between ATFL’s inferior fascicle and CFL transmit tension between both ligaments. Knee Surg. Sport. Traumatol. Arthrosc. 2021, 29, 2511–2516. [Google Scholar] [CrossRef]
- Bonnel, F.; Toullec, E.; Mabit, C.; Tourné, Y. Chronic ankle instability: Biomechanics and pathomechanics of ligaments injury and associated lesions. Orthop. Traumatol. Surg. Res. 2010, 96, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Gribble, P.A.; Delahunt, E.; Bleakley, C.; Caulfield, B.; Docherty, C.; Fourchet, F.; Fong, D.; Hertel, J.; Hiller, C.; Kaminski, T.; et al. Selection criteria for patients with chronic ankle instability in controlled research: A position statement of the International Ankle Consortium. J. Orthop. Sport. Phys. Ther. 2013, 43, 585–591. [Google Scholar] [CrossRef]
- de Souza, R.W.; Silva, D.S.; Passos, L.A.; Roder, M.; Santana, M.C.; Pinheiro, P.R.; de Albuquerque, V.H.C. Computer-assisted Parkinson’s disease diagnosis using fuzzy optimum- path forest and Restricted Boltzmann Machines. Comput. Biol. Med. 2021, 131, 104260. [Google Scholar] [CrossRef] [PubMed]
- Odusami, M.; Maskeliūnas, R.; Damaševičius, R. An Intelligent System for Early Recognition of Alzheimer’s Disease Using Neuroimaging. Sensors 2022, 22, 740. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, Y.; Liang, Y.; Zhan, Z.; Jiang, M.; Zhang, X.; Daniel, S.d.S.; Wu, W.; Albuquerque, V.H.C.d. Edge2Analysis: A Novel AIoT Platform for Atrial Fibrillation Recognition and Detection. IEEE J. Biomed. Health Inform. 2022, 26, 5772–5782. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, M.; Zhang, X.; da Silva, D.S.; de Albuquerque, V.H.C.; Wu, W. Implementing ultra-lightweight co-inference model in ubiquitous edge device for atrial fibrillation detection. Expert Syst. Appl. 2022, 216, 119407. [Google Scholar] [CrossRef]
- Sousa, F.D.O.; da Silva, D.S.; Cavalcante, T.D.S.; Neto, E.C.; Gondim, V.J.T.; Nogueira, I.C.; de Alexandria, A.R.; de Albuquerque, V.H.C. A Novel Virtual Nasal Endoscopy System based on Computed Tomography Scans. Virtual Real. Intell. Hardw. 2022, 4, 359–379. [Google Scholar] [CrossRef]
- de Mesquita, V.A.; Cortez, P.C.; Ribeiro, A.B.; de Albuquerque, V.H.C. A novel method for lung nodule detection in computed tomography scans based on Boolean equations and vector of filters techniques. Comput. Electr. Eng. 2022, 100, 107911. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, G.; Chen, S.; de Albuquerque, V.H.C. An Intelligent Multisampling Tensor Model for Oral Cancer Classification. IEEE Trans. Ind. Inform. 2022, 18, 7853–7861. [Google Scholar] [CrossRef]
- da Silva, D.S.; Nascimento, C.S.; Jagatheesaperumal, S.K.; Albuquerque, V.H.C.d. Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis. Sensors 2022, 22, 8818. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Maskeliūnas, R. TTCNN: A Breast Cancer Detection and Classification towards Computer-Aided Diagnosis Using Digital Mammography in Early Stages. Appl. Sci. 2022, 12, 3273. [Google Scholar] [CrossRef]
- Erdogan, A.; Satici, A.C.; Patoglu, V. Design of a reconfigurable force feedback ankle exoskeleton for physical therapy. In Proceedings of the 2009 ASME/IFToMM International Conference on Reconfigurable Mechanisms and Robots, London, UK, 22–24 June 2009; pp. 400–408. [Google Scholar]
- Barua, A.; Zakia, U.; Menon, C.; Jiang, X. Deep Learning Techniques in Estimating Ankle Joint Power Using Wearable IMUs. IEEE Access 2021, 9, 83041–83054. [Google Scholar] [CrossRef]
- Behboodi, A.; Lee, W.A.; Bulea, T.C.; Damiano, D.L. Evaluation of Multi-layer Perceptron Neural Networks in Predicting Ankle Dorsiflexion in Healthy Adults using Movement-related Cortical Potentials for BCI-Neurofeedback Applications. In Proceedings of the 2022 International Conference on Rehabilitation Robotics (ICORR), Rotterdam, The Netherlands, 25–29 July 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Rebouças Filho, P.P.; Cortez, P.C.; da Silva Barros, A.C.; de Albuquerque, V.H.C. Novel Adaptive Balloon Active Contour Method based on internal force for image segmentation—A systematic evaluation on synthetic and real images. Expert Syst. Appl. 2014, 41, 7707–7721. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, X.; Sun, S.; Wu, D.; Bai, J.; Yin, Y.; Liu, X.; Zhang, H.; de Albuquerque, V.H.C. Learning physical properties in complex visual scenes: An intelligent machine for perceiving blood flow dynamics from static CT angiography imaging. Neural Netw. 2020, 123, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Parah, S.A.; Kaw, J.A.; Bellavista, P.; Loan, N.A.; Bhat, G.M.; Muhammad, K.; de Albuquerque, V.H.C. Efficient Security and Authentication for Edge-Based Internet of Medical Things. IEEE Internet Things J. 2021, 8, 15652–15662. [Google Scholar] [CrossRef]
- Ullah, H.; Muhammad, K.; Irfan, M.; Anwar, S.; Sajjad, M.; Imran, A.S.; de Albuquerque, V.H.C. Light-DehazeNet: A Novel Lightweight CNN Architecture for Single Image Dehazing. IEEE Trans. Image Process. 2021, 30, 8968–8982. [Google Scholar] [CrossRef] [PubMed]
- Doherty, C.; Delahunt, E.; Caulfield, B.; Hertel, J.; Ryan, J.; Bleakley, C. The incidence and prevalence of ankle sprain injury: A systematic review and meta-analysis of prospective epidemiological studies. Sport. Med. 2014, 44, 123–140. [Google Scholar] [CrossRef]
- Raheem, O.A.; O’Brien, M. Anatomical review of the lateral collateral ligaments of the ankle: A cadaveric study. Anat. Sci. Int. 2011, 86, 189–193. [Google Scholar] [CrossRef]
- Clanton, T.O.; Campbell, K.J.; Wilson, K.J.; Michalski, M.P.; Goldsmith, M.T.; Wijdicks, C.A.; LaPrade, R.F. Qualitative and quantitative anatomic investigation of the lateral ankle ligaments for surgical reconstruction procedures. JBJS 2014, 96, e98. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Su, M.; Chen, Z.; Qu, R.; Yuan, Z.; Yuan, J.; He, S.; Li, Z.; Liu, C.; Xiao, Z.; et al. Anatomic Measurement and Variability Analysis of the Anterior Talofibular Ligament and Calcaneofibular Ligament of the Ankle. Orthop. J. Sport. Med. 2021, 9, 23259671211047269. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man, Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Sari, Y.; Baskara, A.R.; Wahyuni, R. Classification of Chili Leaf Disease Using the Gray Level Co-occurrence Matrix (GLCM) and the Support Vector Machine (SVM) Methods. In Proceedings of the 2021 Sixth International Conference on Informatics and Computing (ICIC), Jakarta, Indonesia, 3–4 November 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Nguyen, T.H.; Ngo, B.V. A GLCM Algorithm for Optimal Features of Mammographic Images for Detection of Breast Cancer. In Proceedings of the 2021 International Conference on System Science and Engineering (ICSSE), Ho Chi Minh City, Vietnam, 26–28 August 2021; pp. 295–299. [Google Scholar] [CrossRef]
- Alfa Farah, M.N.; Hayyin Suristiyanti, W.; Ibad, S.; Pramunendar, R.A.; Fajar Shidik, G. GLCM Feature Extraction and PCA for Tuberculosis Detection with Neural Network. In Proceedings of the 2022 International Seminar on Application for Technology of Information and Communication (iSemantic), Semarang, Indonesia, 17–18 September 2022; pp. 314–318. [Google Scholar] [CrossRef]
- Aqreerah, S.; Alariyibi, A.; El-Tarhouni, W. Multispectral palmprint recognition based on three descriptors: LBP, Shift LBP, and Multi Shift LBP with LDA classifier. In Proceedings of the 2022 IEEE 2nd International Maghreb Meeting of the Conference on Sciences and Techniques of Automatic Control and Computer Engineering (MI-STA), Sabratha, Liby, 23–25 May 2022; pp. 506–510. [Google Scholar] [CrossRef]
- Sari, B.P.; Jusman, Y. Classification System for Cervical Cell Images based on Hu Moment Invariants Methods and Support Vector Machine. In Proceedings of the 2021 International Conference on Intelligent Technologies (CONIT), Hubli, India, 25–27 June 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Aravinda, C.; Meng, L.; Uday Kumar Reddy, K.; Prabhu, A. Signature Recognition and Verification Using Multiple Classifiers Combination of Hu’s and HOG Features. In Proceedings of the 2019 International Conference on Advanced Mechatronic Systems (ICAMechS), Kusatsu, Japan, 26–28 August 2019; pp. 63–68. [Google Scholar] [CrossRef]
- Dutta, J.; Chanda, D. Music Emotion Recognition in Assamese Songs using MFCC Features and MLP Classifier. In Proceedings of the 2021 International Conference on Intelligent Technologies (CONIT), Hubli, India, 25–27 June 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Li, D.; Wang, H.; Li, Z. Accurate and Fast Wavelength Demodulation for Fbg Reflected Spectrum Using Multilayer Perceptron (Mlp) Neural Network. In Proceedings of the 2020 12th International Conference on Measuring Technology and Mechatronics Automation (ICMTMA), Phuket, Thailand, 28–29 February 2020; pp. 265–268. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Maskeliūnas, R. Multi-Modal Brain Tumor Detection Using Deep Neural Network and Multiclass SVM. Medicina 2022, 58, 1090. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiao, X.; Li, Y.; Mi, Q.; Yang, Z. Effective Data Classification via Combining Neural Networks and SVM. In Proceedings of the 2019 Chinese Control And Decision Conference (CCDC), Nanchang, China, 3–5 June 2019; pp. 4006–4009. [Google Scholar] [CrossRef]
- Sindhu, S.; Patil, S.P.; Sreevalsan, A.; Rahman, F.; Saritha, A.N. Phishing Detection using Random Forest, SVM and Neural Network with Backpropagation. In Proceedings of the 2020 International Conference on Smart Technologies in Computing, Electrical and Electronics (ICSTCEE), Bengaluru, India, 9–10 October 2020; pp. 391–394. [Google Scholar] [CrossRef]
- Issa, M.E.; Helm, A.M.; Al-Qaness, M.A.A.; Dahou, A.; Elaziz, M.A.; Damaševičius, R. Human Activity Recognition Based on Embedded Sensor Data Fusion for the Internet of Healthcare Things. Healthcare 2022, 10, 1084. [Google Scholar] [CrossRef]
- Xiang, Y.; Li, L.; Zhou, W. Random Forest Classifier For Hardware Trojan Detection. In Proceedings of the 2019 12th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 14–15 December 2019; Volume 1, pp. 134–137. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Y.; Jiang, W. Application of K-Nearest Neighbor (KNN) Algorithm for Human Action Recognition. In Proceedings of the 2021 IEEE 4th Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Chongqing, China, 18–20 June 2021; Volume 4, pp. 492–496. [Google Scholar] [CrossRef]
- Ogundokun, R.O.; Arowolo, M.O.; Misra, S.; Damasevicius, R. An Efficient Blockchain-Based IoT System Using Improved KNN Machine Learning Classifier. In Blockchain based Internet of Things; Lecture Notes on Data Engineering and Communications Technologies; Springer: Singapore, 2022; Volume 112, pp. 171–180. [Google Scholar]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Liu, F.; Zhou, Z.; Samsonov, A.; Blankenbaker, D.; Larison, W.; Kanarek, A.; Lian, K.; Kambhampati, S.; Kijowski, R. Deep learning approach for evaluating knee MR images: Achieving high diagnostic performance for cartilage lesion detection. Radiology 2018, 289, 160. [Google Scholar] [CrossRef]
- Bien, N.; Rajpurkar, P.; Ball, R.L.; Irvin, J.; Park, A.; Jones, E.; Bereket, M.; Patel, B.N.; Yeom, K.W.; Shpanskaya, K.; et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med. 2018, 15, e1002699. [Google Scholar] [CrossRef] [PubMed] [Green Version]


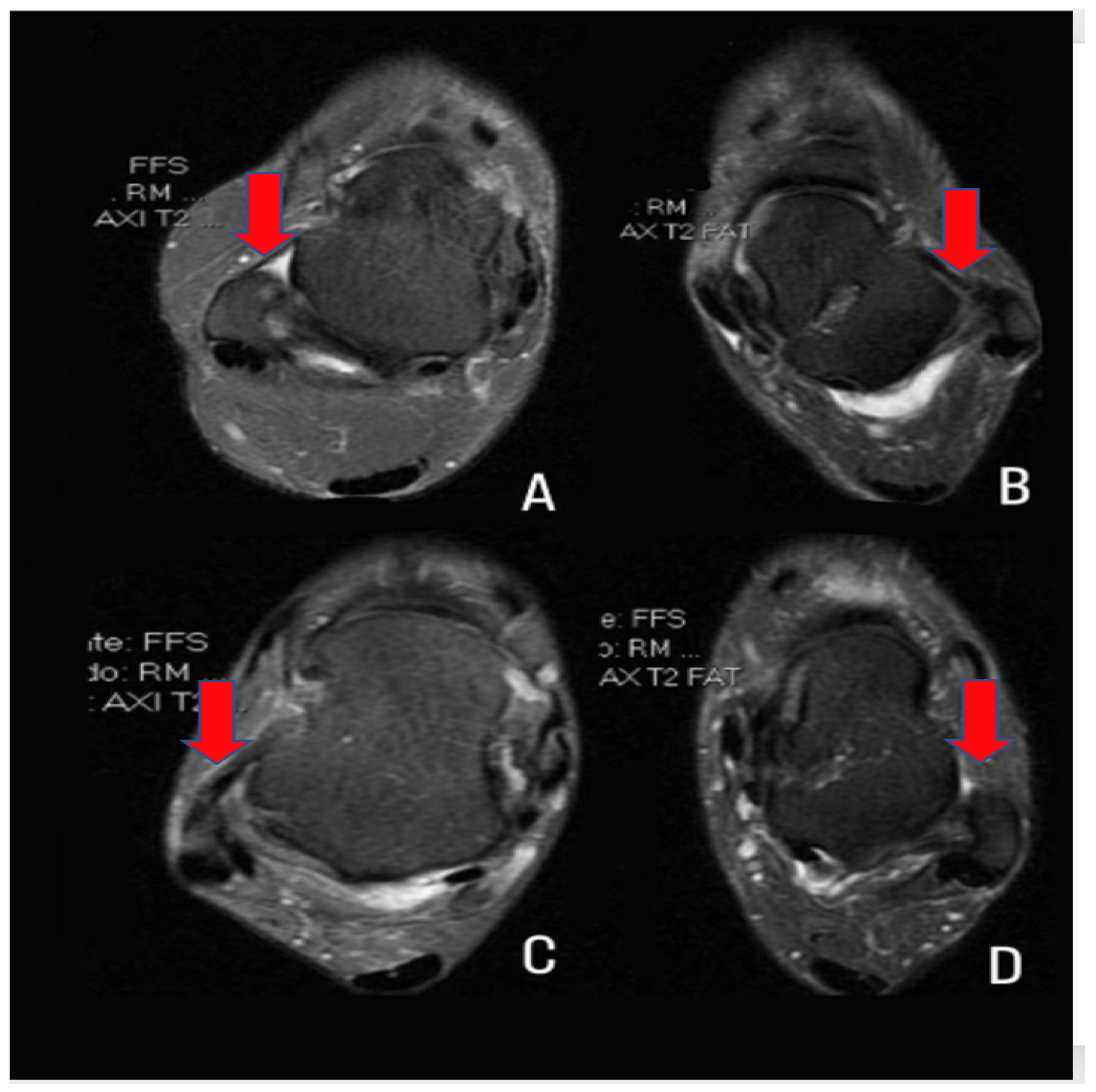







| Standard inclusion criteria endorsed by the international ankle consortium for enrolling patients who fall within the heterogeneous condition of chronic ankle instability in controlled research | |
| 1. A history of at least one significant ankle sprain | |
| At least 12 months prior to study enrollment | Associated with inflammatory symptoms |
| Created at least one interrupted day of desired physical activity | Acute traumatic injury to the lateral ligament complex of the ankle joint as a result of excessive inversion of the rear foot or a combined plantar flexion and adduction of the foot. This usually results in some initial deficits of functional and disability. |
| 2. A history of the previously injured ankle joint “giving way”, and/or recurrent sprain, and/or “feelings of instability” | |
| Subjects should report at least two episodes of giving way in the six months prior to study enrollment | Giving way: the recurring occurrence of uncontrolled and unpredictable bouts of excessive rear foot inversion that do not result in an acute lateral ankle injury. |
| Recurrent sprain: two or more sprains to the same ankle | Self-reported ankle instability confirmed with a validate ankle instability, a specific questionnaire using the associated cutoff score: Ankle Instability Instrument, answering yes to at least five yes/no question. |
| 3. Foot and Ankle Outcome Score: score of <75% in three or more categories | |
| Metric Values, % | Normal Ligament | Absent Ligament | Abnormal Ligament |
|---|---|---|---|
| Sensitivity | 100% | 68% | 100% |
| Specificity | 16% | 63% | 22% |
| Accuracy | 26% | 63% | 16% |
| Algorithm | Time (s) |
|---|---|
| GLCM | 2.219 |
| LBP | 0.121 |
| HU | 0.026 |
| DC | 1.528 |
| Algorithms | Representation |
|---|---|
| GLCM | Set 1 |
| LBP | Set 2 |
| HU | Set 3 |
| DC | Set 4 |
| GLCM + LBP | Set 5 |
| GLCM + HU | Set 6 |
| GLCM + DC | Set 7 |
| LBP + HU | Set 8 |
| LBP + DC | Set 9 |
| HU + DC | Set 10 |
| GLCM + HU + LBP | Set 11 |
| GLCM + DC + LBP | Set 12 |
| GLCM + HU + DC | Set 13 |
| LBP + HU + DC | Set 14 |
| GLCM + LBP + HU + DC | Set 15 |
| Algorithms | Metrics (%) | MLP | kNN | SVM | RF |
|---|---|---|---|---|---|
| Set 1 | ACC Global | 49.26 ± 2.12 | 55.73 ± 5.18 | 59.75 ± 5.14 | 70.60 ± 5.74 |
| ATFL | 49.40 ± 43.60 | 54.70 ± 11.94 | 57.64 ± 13.49 | 75.98 ± 11.03 | |
| HealthyControl | 48.92 ± 44.98 | 56.79 ± 11.66 | 61.83 ± 14.96 | 65.29 ± 10.60 | |
| F1-score | 35.99 ± 30.36 | 54.62 ± 7.35 | 58.13 ± 7.06 | 96.50 ± 1.82 | |
| Set 2 | ACC Global | 74.87 ± 3.94 | 72.56 ± 7.51 | 58.90 ± 5.88 | 80.60 ± 4.84 |
| ATFL | 73.28 ± 8.95 | 72.20 ± 12.38 | 81.69 ± 15.37 | 83.35 ± 6.98 | |
| HealthyControl | 76.39 ± 8.20 | 72.69 ± 10.19 | 36.94 ± 22.44 | 77.72 ± 9.70 | |
| F1-score | 74.21 ± 4.75 | 72.03 ± 8.65 | 66.05 ± 4.75 | 81.11 ± 4.45 | |
| Set 3 | ACC Global | 48.78 ± 1.11 | 44.51 ± 6.06 | 48.78 ± 1.11 | 51.58 ± 7.28 |
| ATFL | 60.00 ± 48.98 | 43.40 ± 9.06 | 60.00 ± 48.98 | 49.78 ± 9.62 | |
| HealthyControl | 40.00 ± 48.98 | 45.69 ± 8.10 | 40.00 ± 48.98 | 53.36 ± 9.57 | |
| F1-score | 39.34 ± 32.12 | 43.49 ± 7.72 | 39.34 ± 32.12 | 50.35 ± 7.92 | |
| Set 4 | ACC Global | 62.80 ± 6.40 | 60.36 ± 5.91 | 65.85 ± 5.82 | 65.73 ± 6.38 |
| ATFL | 65.55 ± 12.84 | 61.82 ± 11.46 | 77.98 ± 14.22 | 68.23 ± 13.68 | |
| HealthyControl | 60.19 ± 14.29 | 58.92 ± 8.96 | 53.70 ± 12.01 | 63.27 ± 9.19 | |
| F1-score | 63.32 ± 7.54 | 60.47 ± 7.72 | 69.00 ± 6.98 | 65.94 ± 8.38 | |
| Set 5 | ACC Global | 51.82 ± 4.74 | 58.29 ± 6.16 | 64.75 ± 6.09 | 81.95 ± 4.52 |
| ATFL | 30.88 ± 38.94 | 54.96 ± 12.33 | 69.63 ± 10.70 | 85.03 ± 7.10 | |
| HealthyControl | 72.67 ± 38.65 | 61.72 ± 9.15 | 59.98 ± 10.15 | 78.92 ± 7.65 | |
| F1-score | 26.37 ± 28.20 | 56.11 ± 8.96 | 66.02 ± 6.53 | 82.35 ± 4.54 | |
| Set 6 | ACC Global | 53.04 ± 5.71 | 55.48 ± 6.97 | 65.24 ± 6.97 | 71.09 ± 4.89 |
| ATFL | 17.95 ± 29.87 | 54.64 ± 12.58 | 70.03 ± 13.38 | 76.54 ± 8.30 | |
| HealthyControl | 87.30 ± 23.85 | 56.50 ± 10.91 | 60.35 ± 9.23 | 65.74 ± 8.65 | |
| F1-score | 17.99 ± 25.98 | 54.52 ± 8.65 | 66.25 ± 8.80 | 72.43 ± 5.06 | |
| Set 7 | ACC Global | 50.36 ± 8.70 | 59.51 ± 5.99 | 68.65 ± 7.95 | 76.46 ± 6.51 |
| ATFL | 68.92 ± 30.36 | 60.99 ± 12.56 | 72.57 ± 14.54 | 80.97 ± 9.90 | |
| HealthyControl | 32.15 ± 34.19 | 57.95 ± 10.00 | 64.82 ± 10.35 | 71.84 ± 7.67 | |
| F1-score | 53.64 ± 20.43 | 59.63 ± 7.93 | 69.31 ± 9.70 | 77.34 ± 6.88 | |
| Set 8 | ACC Global | 71.95 ± 6.83 | 68.41 ± 9.64 | 58.04 ± 6.00 | 78.78 ± 5.92 |
| ATFL | 73.58 ± 9.03 | 67.66 ± 11.93 | 70.89 ± 29.58 | 80.19 ± 8.07 | |
| HealthyControl | 70.41 ± 10.63 | 69.21 ± 10.96 | 46.39 ± 26.99 | 77.44 ± 9.46 | |
| F1-score | 72.25 ± 6.58 | 67.88 ± 10.01 | 58.24 ± 20.06 | 78.96 ± 5.77 | |
| Set 9 | ACC Global | 65.12 ± 5.97 | 59.39 ± 5.41 | 58.04 ± 6.00 | 62.80 ± 4.21 |
| ATFL | 69.96 ± 14.58 | 63.65 ± 9.00 | 70.89 ± 29.58 | 75.02 ± 12.15 | |
| HealthyControl | 60.27 ± 10.47 | 55.17 ± 9.08 | 46.39 ± 26.99 | 50.88 ± 13.31 | |
| F1-score | 65.89 ± 8.20 | 60.68 ± 5.89 | 58.24 ± 20.06 | 66.36 ± 4.80 | |
| Set 10 | ACC Global | 60.12 ± 6.78 | 57.92 ± 4.99 | 65.48 ± 6.13 | 64.63 ± 4.11 |
| ATFL | 61.95 ± 14.54 | 59.32 ± 10.99 | 77.72 ± 12.80 | 69.15 ± 10.49 | |
| HealthyControl | 58.48 ± 9.81 | 56.55 ± 7.64 | 53.39 ± 12.50 | 60.21 ± 9.55 | |
| F1-score | 60.07 ± 10.43 | 58.13 ± 7.06 | 68.98 ± 6.47 | 65.87 ± 5.56 | |
| Set 11 | ACC Global | 49.39 ± 1.86 | 58.17 ± 6.95 | 62.31 ± 4.39 | 78.90 ± 5.93 |
| ATFL | 9.25 ± 27.76 | 55.41 ± 9.15 | 69.30 ± 7.65 | 82.65 ± 8.35 | |
| HealthyControl | 90.00 ± 30.00 | 61.01 ± 13.41 | 55.30 ± 9.33 | 75.13 ± 7.91 | |
| F1-score | 6.21 ± 18.65 | 56.86 ± 6.71 | 64.68 ± 4.11 | 79.55 ± 6.11 | |
| Set 12 | ACC Global | 52.56 ± 5.89 | 60.85 ± 6.38 | 71.09 ± 7.03 | 80.60 ± 5.08 |
| ATFL | 42.41 ± 43.44 | 59.57 ± 9.06 | 75.77 ± 12.73 | 84.34 ± 6.79 | |
| HealthyControl | 62.39 ± 42.59 | 62.23 ± 10.82 | 66.39 ± 9.33 | 76.83 ± 8.99 | |
| F1-score | 33.87 ± 31.76 | 60.48 ± 6.49 | 64.68 ± 4.11 | 81.48 ± 4.68 | |
| Set 13 | ACC Global | 50.60 ± 4.28 | 62.19 ± 5.94 | 70.36 ± 6.95 | 76.09 ± 6.43 |
| ATFL | 68.67 ± 37.71 | 61.95 ± 9.41 | 75.64 ± 9.73 | 81.19 ± 8.34 | |
| HealthyControl | 33.40 ± 39.97 | 62.48 ± 9.89 | 65.30 ± 13.51 | 71.00 ± 7.47 | |
| F1-score | 50.97 ± 23.54 | 61.89 ± 6.58 | 71.79 ± 6.31 | 77.15 ± 6.33 | |
| Set 14 | ACC Global | 53.53 ± 7.99 | 60.24 ± 6.41 | 67.92 ± 5.57 | 82.56 ± 6.27 |
| ATFL | 39.85 ± 40.03 | 60.75 ± 9.12 | 76.44 ± 6.62 | 83.85 ± 8.96 | |
| HealthyControl | 68.01 ± 38.75 | 59.72 ± 8.70 | 59.53 ± 10.73 | 81.36 ± 8.70 | |
| F1-score | 34.45 ± 27.98 | 60.11 ± 6.70 | 70.31 ± 4.53 | 82.62 ± 6.43 | |
| Set 15 | ACC Global | 56.09 ± 10.34 | 60.48 ± 6.96 | 72.43 ± 5.29 | 81.70 ± 5.94 |
| ATFL | 49.59 ± 31.67 | 61.88 ± 12.69 | 78.58 ± 13.05 | 83.38 ± 11.25 | |
| HealthyControl | 63.27 ± 32.64 | 58.98 ± 12.01 | 66.35 ± 8.56 | 79.95 ± 9.35 | |
| F1-score | 46.76 ± 24.75 | 60.55 ± 8.49 | 73.54 ± 7.02 | 81.77 ± 6.47 |
| Algorithms | Metrics (%) | MLP | kNN | SVM | RF |
|---|---|---|---|---|---|
| Set 1 | Training | 0.01696 | 0.00026 | 7.99108 | 0.07679 |
| Test | 0.00024 | 0.00110 | 0.00022 | 0.00659 | |
| Set 2 | Training | 0.78163 | 0.00019 | 0.00087 | 0.08279 |
| Test | 0.00025 | 0.00109 | 0.00019 | 0.00672 | |
| Set 3 | Training | 0.01699 | 0.00031 | 0.00090 | 0.08290 |
| Test | 0.00028 | 0.00115 | 0.00024 | 0.00718 | |
| Set 4 | Training | 1.01803 | 0.00031 | 0.13495 | 0.07867 |
| Test | 0.00025 | 0.00115 | 0.00022 | 0.00683 | |
| Set 5 | Training | 0.03747 | 0.00020 | 3.61693 | 0.07941 |
| Test | 0.00023 | 0.00099 | 0.00023 | 0.00672 | |
| Set 6 | Training | 0.02615 | 0.00030 | 3.67280 | 0.08182 |
| Test | 0.00026 | 0.00112 | 0.00024 | 0.00696 | |
| Set 7 | Training | 0.03339 | 0.00029 | 13.56860 | 0.08361 |
| Test | 0.00026 | 0.00121 | 0.00021 | 0.00690 | |
| Set 8 | Training | 0.90728 | 0.00020 | 0.00093 | 0.08563 |
| Test | 0.00022 | 0.00108 | 0.00021 | 0.00696 | |
| Set 9 | Training | 0.77169 | 0.00021 | 0.14240 | 0.08291 |
| Test | 0.00029 | 0.00107 | 0.00024 | 0.00679 | |
| Set 10 | Training | 1.41660 | 0.00028 | 0.12939 | 0.08386 |
| Test | 0.00027 | 0.00121 | 0.00023 | 0.00743 | |
| Set 11 | Training | 0.02186 | 0.00020 | 3.24591 | 0.08495 |
| Test | 0.00023 | 0.00109 | 0.00023 | 0.00688 | |
| Set 12 | Training | 0.03380 | 0.00019 | 11.99871 | 0.08607 |
| Test | 0.00027 | 0.00118 | 0.00022 | 0.00666 | |
| Set 13 | Training | 0.03712 | 0.00018 | 13.16548 | 0.07790 |
| Test | 0.00024 | 0.00105 | 0.00023 | 0.00662 | |
| Set 14 | Training | 0.04402 | 0.00021 | 14.03406 | 0.08386 |
| Test | 0.00027 | 0.00105 | 0.00023 | 0.00665 | |
| Set 15 | Training | 0.08085 | 0.00022 | 12.39197 | 0.08032 |
| Test | 0.00029 | 0.00106 | 0.00023 | 0.00638 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Astolfi, R.S.; da Silva, D.S.; Guedes, I.S.; Nascimento, C.S.; Damaševičius, R.; Jagatheesaperumal, S.K.; de Albuquerque, V.H.C.; Leite, J.A.D. Computer-Aided Ankle Ligament Injury Diagnosis from Magnetic Resonance Images Using Machine Learning Techniques. Sensors 2023, 23, 1565. https://doi.org/10.3390/s23031565
Astolfi RS, da Silva DS, Guedes IS, Nascimento CS, Damaševičius R, Jagatheesaperumal SK, de Albuquerque VHC, Leite JAD. Computer-Aided Ankle Ligament Injury Diagnosis from Magnetic Resonance Images Using Machine Learning Techniques. Sensors. 2023; 23(3):1565. https://doi.org/10.3390/s23031565
Chicago/Turabian StyleAstolfi, Rodrigo S., Daniel S. da Silva, Ingrid S. Guedes, Caio S. Nascimento, Robertas Damaševičius, Senthil K. Jagatheesaperumal, Victor Hugo C. de Albuquerque, and José Alberto D. Leite. 2023. "Computer-Aided Ankle Ligament Injury Diagnosis from Magnetic Resonance Images Using Machine Learning Techniques" Sensors 23, no. 3: 1565. https://doi.org/10.3390/s23031565
APA StyleAstolfi, R. S., da Silva, D. S., Guedes, I. S., Nascimento, C. S., Damaševičius, R., Jagatheesaperumal, S. K., de Albuquerque, V. H. C., & Leite, J. A. D. (2023). Computer-Aided Ankle Ligament Injury Diagnosis from Magnetic Resonance Images Using Machine Learning Techniques. Sensors, 23(3), 1565. https://doi.org/10.3390/s23031565








