Energy-Efficient EEG-Based Scheme for Autism Spectrum Disorder Detection Using Wearable Sensors
Abstract
1. Introduction
2. Design of the ASD Detection Scheme
2.1. The Proposed Approach for ASD Detection
2.2. Signal Analysis and Feature Extraction
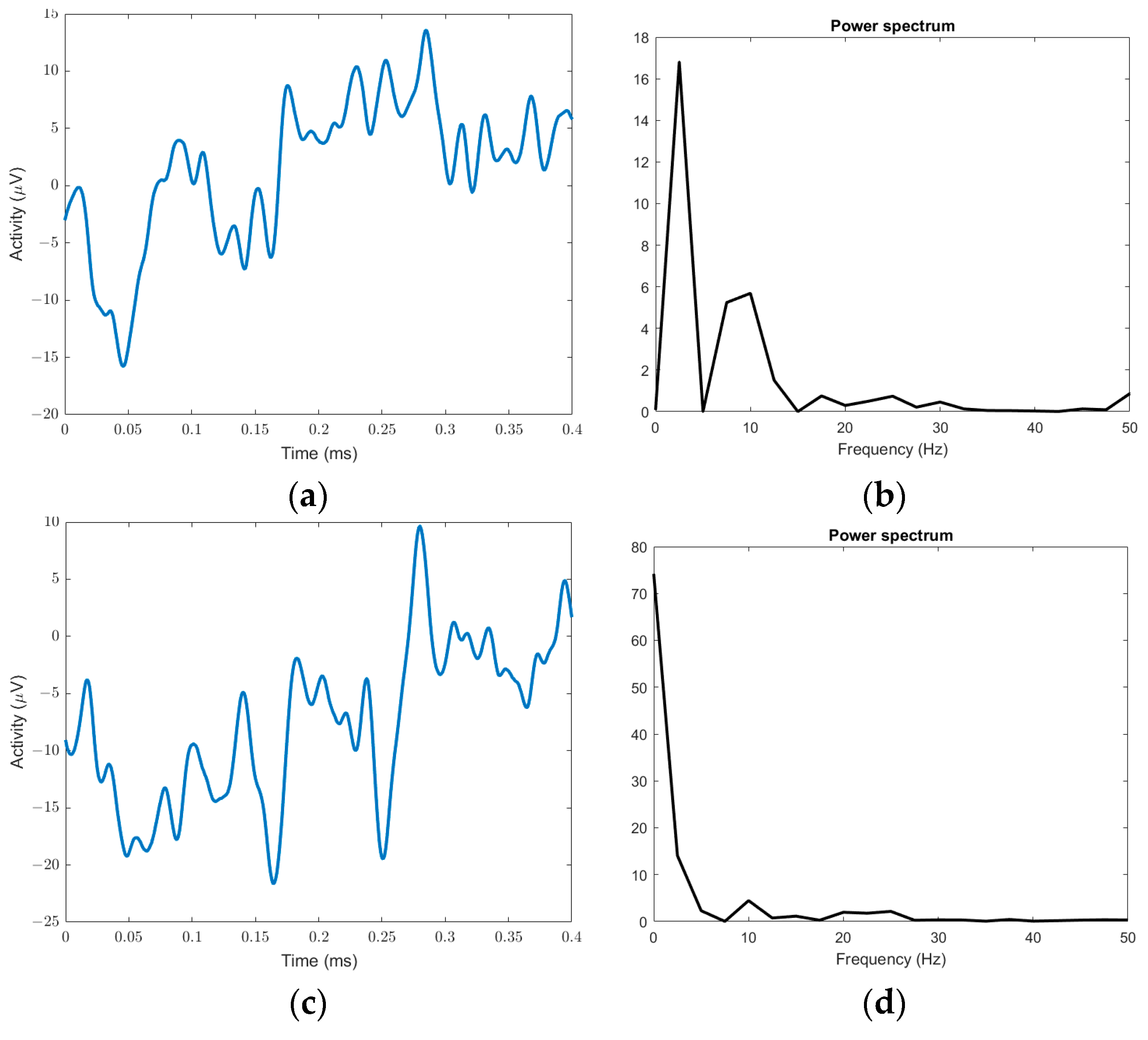
2.2.1. Signal Transform
2.2.2. Feature Extraction
2.3. Feature Selection
2.4. Classification and ASD Detection
2.4.1. Support Vector Machine
2.4.2. Logistic Regression
2.4.3. Decision Tree
3. Performances Evaluation of the Proposed Scheme
3.1. Classification of ASD Cases
- Accuracy (Acc): It provides the correct prediction of the classifier.
- Sensitivity or recall (Sen): It expresses the ability of the scheme to identify subjects who have ASD correctly.
- Specificity (Spec): It shows the scheme’s ability to identify typical developing subjects correctly.
- Positive Predictive Value (PPV) or Precision: It provides the probability of how likely it is that the subject has ASD.
- Negative Predictive Value (NPV): It expresses the probability of how likely it is that the subject is a typical developing subject.
- F1-score (F1): It combines both sensitivity and PPV in a single metric.
3.2. Energy-Consumption Estimation of the Proposed Scheme
- On-node feature extraction and classification: In this scenario, we evaluated the energy consumption related to the processing of the EEG signal and the extraction of the features and the classification at the wearable sensor. We implemented the process related to the extraction of the features that provided the highest accuracy, 96%, in our scheme (Table 6).
- ∘
- For the classification with SVM and logistic regression, the EEG signal was processed in the gamma sub-band. We evaluated the deployment of the best performance features (absolute Welch, ApEn, and ApEn normalized). For the classification, we added the decision classification Equation (11) for SVM and Equation (12) for logistic regression.
- ∘
- For the classification with the decision tree algorithm, the EEG signal was processed in the alpha sub-band. The energy consumption was evaluated for four features of the proposed scheme (absolute Welch, relative Welch, variance, and ApEn normalized). For the classification, we have added the if-else rules resulting from the decision tree model.
- Streaming raw EEG signal segment: This scenario is based on the idea of streaming raw EEG signal as in the traditional computerized scheme.
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- CDC Data and Statistics on Autism Spectrum Disorder|CDC. Available online: https://www.cdc.gov/ncbddd/autism/data.html (accessed on 30 May 2022).
- Jeste, S.S.; Frohlich, J.; Loo, S.K. Electrophysiological Biomarkers of Diagnosis and Outcome in Neurodevelopmental Disorders. Curr. Opin. Neurol. 2015, 28, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Autism and Autism Spectrum Disorders. Available online: https://www.apa.org/topics/autism-spectrum-disorder (accessed on 29 May 2022).
- Elder, J.; Kreider, C.; Brasher, S.; Ansell, M. Clinical Impact of Early Diagnosis of Autism on the Prognosis and Parent-Child Relationships. PRBM 2017, 10, 283–292. [Google Scholar] [CrossRef]
- Gabard-Durnam, L.J.; Wilkinson, C.; Kapur, K.; Tager-Flusberg, H.; Levin, A.R.; Nelson, C.A. Longitudinal EEG Power in the First Postnatal Year Differentiates Autism Outcomes. Nat. Commun. 2019, 10, 4188. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, R.C.; Maye, M.P.; Carter, A.S. Age at First Identification of Autism Spectrum Disorder: An Analysis of Two US Surveys. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Bosl, W.J.; Tager-Flusberg, H.; Nelson, C.A. EEG Analytics for Early Detection of Autism Spectrum Disorder: A Data-Driven Approach. Sci. Rep. 2018, 8, 6828. [Google Scholar] [CrossRef]
- Brihadiswaran, G.; Haputhanthri, D.; Gunathilaka, S.; Meedeniya, D.; Jayarathna, S. EEG-Based Processing and Classification Methodologies for Autism Spectrum Disorder: A Review. J. Comput. Sci. 2019, 15, 1161–1183. [Google Scholar] [CrossRef]
- Gurau, O.; Bosl, W.J.; Newton, C.R. How Useful Is Electroencephalography in the Diagnosis of Autism Spectrum Disorders and the Delineation of Subtypes: A Systematic Review. Front. Psychiatry 2017, 8, 121. [Google Scholar] [CrossRef]
- Bhat, S.; Acharya, U.R.; Adeli, H.; Bairy, G.M.; Adeli, A. Automated Diagnosis of Autism: In Search of a Mathematical Marker. Rev. Neurosci. 2014, 25, 851–861. [Google Scholar] [CrossRef]
- Reiersen, A.M. Early Identification of Autism Spectrum Disorder: Is Diagnosis by Age 3 a Reasonable Goal? J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 284–285. [Google Scholar] [CrossRef]
- Billeci, L.; Sicca, F.; Maharatna, K.; Apicella, F.; Narzisi, A.; Campatelli, G.; Calderoni, S.; Pioggia, G.; Muratori, F. On the Application of Quantitative EEG for Characterizing Autistic Brain: A Systematic Review. Front. Hum. Neurosci. 2013, 7, 442. [Google Scholar] [CrossRef]
- Siuly, S.; Li, Y.; Zhang, Y. EEG Signal Analysis and Classification; Health Information Science; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-47652-0. [Google Scholar]
- Joshi, V.; Nanavati, N. A Review of EEG Signal Analysis for Diagnosis of Neurological Disorders Using Machine Learning. J.-BPE 2021, 7, 040201. [Google Scholar] [CrossRef]
- Heunis, T.; Aldrich, C.; Peters, J.M.; Jeste, S.S.; Sahin, M.; Scheffer, C.; de Vries, P.J. Recurrence Quantification Analysis of Resting State EEG Signals in Autism Spectrum Disorder—A Systematic Methodological Exploration of Technical and Demographic Confounders in the Search for Biomarkers. BMC Med. 2018, 16, 101. [Google Scholar] [CrossRef]
- Ahmadlou, M.; Adeli, H. Electroencephalograms in Diagnosis of Autism. In Comprehensive Guide to Autism; Patel, V.B., Preedy, V.R., Martin, C.R., Eds.; Springer New York: New York, NY, USA, 2014; pp. 327–343. ISBN 978-1-4614-4787-0. [Google Scholar]
- McPartland, J.C.; Lerner, M.D.; Bhat, A.; Clarkson, T.; Jack, A.; Koohsari, S.; Matuskey, D.; McQuaid, G.A.; Su, W.-C.; Trevisan, D.A. Looking Back at the Next 40 Years of ASD Neuroscience Research. J. Autism. Dev. Disord. 2021, 51, 4333–4353. [Google Scholar] [CrossRef]
- Haartsen, R.; Jones, E.J.H.; Orekhova, E.V.; Charman, T.; Johnson, M.H. The BASIS team. Functional EEG Connectivity in Infants Associates with Later Restricted and Repetitive Behaviours in Autism; a Replication Study. Transl. Psychiatry 2019, 9, 66. [Google Scholar] [CrossRef]
- Orekhova, E.V.; Elsabbagh, M.; Jones, E.J.; Dawson, G.; Charman, T.; Johnson, M.H.; The BASIS Team. EEG Hyper-Connectivity in High-Risk Infants Is Associated with Later Autism. J. Neurodevelop. Disord. 2014, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, A.; Daniel, M.; Marin, A.; Gaonkar, B.; Dapretto, M.; McDonald, N.; Jeste, S. Multivariate Neural Connectivity Patterns in Early Infancy Predict Later Autism Symptoms. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2020, 6, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Song, J.; Li, X.; Kang, J. A Study on EEG Feature Extraction and Classification in Autistic Children Based on Singular Spectrum Analysis Method. Brain Behav. 2020, 10, e01721. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.L.; Levin, A.R.; Gabard-Durnam, L.J.; Tager-Flusberg, H.; Nelson, C.A. Reduced Frontal Gamma Power at 24 Months Is Associated with Better Expressive Language in Toddlers at Risk for Autism. Autism Res. 2019, 12, 1211–1224. [Google Scholar] [CrossRef]
- Heunis, T.-M.; Aldrich, C.; de Vries, P.J. Recent Advances in Resting-State Electroencephalography Biomarkers for Autism Spectrum Disorder—A Review of Methodological and Clinical Challenges. Pediatr. Neurol. 2016, 61, 28–37. [Google Scholar] [CrossRef]
- Lau-Zhu, A.; Lau, M.P.H.; McLoughlin, G. Mobile EEG in Research on Neurodevelopmental Disorders: Opportunities and Challenges. Dev. Cogn. Neurosci. 2019, 36, 100635. [Google Scholar] [CrossRef]
- Ratti, E.; Waninger, S.; Berka, C.; Ruffini, G.; Verma, A. Comparison of Medical and Consumer Wireless EEG Systems for Use in Clinical Trials. Front. Hum. Neurosci. 2017, 11, 398. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovic, V.; Grundlehner, B.; Vullers, R.; Penders, J. Wearable, Wireless EEG Solutions in Daily Life Applications: What Are We Missing? IEEE J. Biomed. Health Inform. 2015, 19, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Manickam, P.; Mariappan, S.A.; Murugesan, S.M.; Hansda, S.; Kaushik, A.; Shinde, R.; Thipperudraswamy, S.P. Artificial Intelligence (AI) and Internet of Medical Things (IoMT) Assisted Biomedical Systems for Intelligent Healthcare. Biosensors 2022, 12, 562. [Google Scholar] [CrossRef]
- Johnson, K.T.; Picard, R.W. Advancing Neuroscience through Wearable Devices. Neuron 2020, 108, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Al-awlaqi, M.A.A.H.; Li, M.; O’Grady, M.; Gu, X.; Wang, J.; Cao, N. Wearable IoT Enabled Real-Time Health Monitoring System. J. Wirel. Com. Netw. 2018, 2018, 298. [Google Scholar] [CrossRef]
- Almusallam, M.; Soudani, A. Feature-Based ECG Sensing Scheme for Energy Efficiency in WBSN. In Proceedings of the 2017 International Conference on Informatics, Health & Technology (ICIHT), Riyadh, Saudi Arabia, 21–23 February 2017; IEEE: Piscataway, NJ, USA; pp. 1–6. [Google Scholar]
- Soudani, A.; Almusallam, M. Atrial Fibrillation Detection Based on ECG-Features Extraction in WBSN. Procedia Comput. Sci. 2018, 130, 472–479. [Google Scholar] [CrossRef]
- Dufort y Alvarez, G.; Favaro, F.; Lecumberry, F.; Martin, A.; Oliver, J.P.; Oreggioni, J.; Ramirez, I.; Seroussi, G.; Steinfeld, L. Wireless EEG System Achieving High Throughput and Reduced Energy Consumption Through Lossless and Near-Lossless Compression. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 231–241. [Google Scholar] [CrossRef]
- Ajani, T.S.; Imoize, A.L.; Atayero, A.A. An Overview of Machine Learning within Embedded and Mobile Devices–Optimizations and Applications. Sensors 2021, 21, 4412. [Google Scholar] [CrossRef]
- Hashemian, M.; Pourghassem, H. Diagnosing Autism Spectrum Disorders Based on EEG Analysis: A Survey. Neurophysiology 2014, 46, 183–195. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, Z. (Eds.) EEG Signal Processing and Feature Extraction; Springer: Singapore, 2019; ISBN 9789811391132. [Google Scholar]
- Al-Fahoum, A.S.; Al-Fraihat, A.A. Methods of EEG Signal Features Extraction Using Linear Analysis in Frequency and Time-Frequency Domains. ISRN Neurosci. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Iftikhar, M.; Khan, S.A.; Hassan, A. A Survey of Deep Learning and Traditional Approaches for EEG Signal Processing and Classification. In Proceedings of the 2018 IEEE 9th Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON), Vancouver, BC, Canada, 1–3 November 2018; p. 6. [Google Scholar]
- Djemal, R.; AlSharabi, K.; Ibrahim, S.; Alsuwailem, A. EEG-Based Computer Aided Diagnosis of Autism Spectrum Disorder Using Wavelet, Entropy, and ANN. BioMed. Res. Int. 2017, 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mallat, S.G. A Wavelet Tour of Signal Processing: The Sparse Way, 3rd ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Boston, MA, USA, 2009; ISBN 978-0-12-374370-1. [Google Scholar]
- Van Drongelen, W. Wavelet Analysis. In Signal Processing for Neuroscientists; Elsevier: Amsterdam, The Netherlands, 2018; pp. 401–423. ISBN 978-0-12-810482-8. [Google Scholar]
- Alhassan, S.; AlDammas, M.A.; Soudani, A. Energy-Efficient Sensor-Based EEG Features’ Extraction for Epilepsy Detection. Procedia Comput. Sci. 2019, 160, 273–280. [Google Scholar] [CrossRef]
- Jiang, X.; Bian, G.-B.; Tian, Z. Removal of Artifacts from EEG Signals: A Review. Sensors 2019, 19, 987. [Google Scholar] [CrossRef]
- Walczak, T.S.; Chokroverty, S. Electroencephalography, Electromyography, and Electro-Oculography: General Principles and Basic Technology. In Sleep Disorders Medicine; Sudhansu, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 157–181. ISBN 978-0-7506-7584-0. [Google Scholar]
- Van Drongelen, W. Continuous, Discrete, and Fast Fourier Transform. In Signal Processing for Neuroscientists; Elsevier: Amsterdam, The Netherlands, 2018; pp. 103–118. ISBN 978-0-12-810482-8. [Google Scholar]
- Sridevi, S.; ShinyDuela, D.J. A comprehensive study on eeg signal processing—methods, challenges and applications. IT in Industry 2021, 9, 3. [Google Scholar]
- Gabard-Durnam, L.; Tierney, A.L.; Vogel-Farley, V.; Tager-Flusberg, H.; Nelson, C.A. Alpha Asymmetry in Infants at Risk for Autism Spectrum Disorders. J. Autism. Dev. Disord. 2015, 45, 473–480. [Google Scholar] [CrossRef]
- Levin, A.R.; Varcin, K.J.; O’Leary, H.M.; Tager-Flusberg, H.; Nelson, C.A. EEG Power at 3 Months in Infants at High Familial Risk for Autism. J. Neurodev. Disord. 2017, 9, 34. [Google Scholar] [CrossRef]
- Damiano-Goodwin, C.R.; Woynaroski, T.G.; Simon, D.M.; Ibañez, L.V.; Murias, M.; Kirby, A.; Newsom, C.R.; Wallace, M.T.; Stone, W.L.; Cascio, C.J. Developmental Sequelae and Neurophysiologic Substrates of Sensory Seeking in Infant Siblings of Children with Autism Spectrum Disorder. Dev. Cogn. Neurosci. 2018, 29, 41–53. [Google Scholar] [CrossRef]
- Simon, D.M.; Damiano, C.R.; Woynaroski, T.G.; Ibañez, L.V.; Murias, M.; Stone, W.L.; Wallace, M.T.; Cascio, C.J. Neural Correlates of Sensory Hyporesponsiveness in Toddlers at High Risk for Autism Spectrum Disorder. J. Autism. Dev. Disord. 2017, 47, 2710–2722. [Google Scholar] [CrossRef]
- Wang, J.; Barstein, J.; Ethridge, L.E.; Mosconi, M.W.; Takarae, Y.; Sweeney, J.A. Resting State EEG Abnormalities in Autism Spectrum Disorders. J. Neurodev. Disord. 2013, 5, 24. [Google Scholar] [CrossRef]
- Van Drongelen, W. 1-D and 2-D Fourier Transform Applications. In Signal Processing for Neuroscientists; Elsevier: Amsterdam, The Netherlands, 2018; pp. 119–152. ISBN 978-0-12-810482-8. [Google Scholar]
- Bhuvaneswari, P.; Kumar, J.S. Influence of Linear Features in Nonlinear Electroencephalography (EEG) Signals. Procedia Comput. Sci. 2015, 47, 229–236. [Google Scholar] [CrossRef]
- Maximo, J.O.; Nelson, C.M.; Kana, R.K. Unrest While Resting? Brain Entropy in Autism Spectrum Disorder. Brain Res. 2021, 1762, 147435. [Google Scholar] [CrossRef]
- Pan, Y.-H.; Lin, W.-Y.; Wang, Y.-H.; Lee, K.-T. Computing multiscale entropy with orthogonal range search. J. Mar. Sci. Technol. 2011, 19, 7. [Google Scholar] [CrossRef]
- Xie, H.-B.; He, W.-X.; Liu, H. Measuring Time Series Regularity Using Nonlinear Similarity-Based Sample Entropy. Phys. Lett. A 2008, 372, 7140–7146. [Google Scholar] [CrossRef]
- Bonnini, S.; Corain, L.; Marozzi, M.; Salmaso, L. Nonparametric Hypothesis Testing: Rank and Permutation Methods with Applications in R; Wiley: Hoboken, NJ, USA, 2014; ISBN 978-1-119-95237-4. [Google Scholar]
- Dodge, Y. The Concise Encyclopedia of Statistics, 1st ed.; Springer: New York, NY, USA, 2008; ISBN 978-0-387-31742-7. [Google Scholar]
- García, S.; Luengo, J.; Herrera, F. Data Preprocessing in Data Mining; Intelligent Systems Reference Library; Springer International Publishing: Cham, Switzerland, 2015; Volume 72, ISBN 978-3-319-10246-7. [Google Scholar]
- Aggarwal, C.C. (Ed.) Data ClassifiCation Algorithms and Applications, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014; ISBN 978-1-4665-8674-1. [Google Scholar]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A Survey on Evolutionary Computation Approaches to Feature Selection. IEEE Trans. Evol. Computat. 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Segato, A.; Marzullo, A.; Calimeri, F.; De Momi, E. Artificial Intelligence for Brain Diseases: A Systematic Review. APL Bioeng. 2020, 4, 041503. [Google Scholar] [CrossRef] [PubMed]
- Koteluk, O.; Wartecki, A.; Mazurek, S.; Kołodziejczak, I.; Mackiewicz, A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. JPM 2021, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Mazlan, A.U.; Sahabudin, N.A.; Remli, M.A.; Ismail, N.S.N.; Mohamad, M.S.; Nies, H.W.; Abd Warif, N.B. A Review on Recent Progress in Machine Learning and Deep Learning Methods for Cancer Classification on Gene Expression Data. Processes 2021, 9, 1466. [Google Scholar] [CrossRef]
- Gupta, C.; Chandrashekar, P.; Jin, T.; He, C.; Khullar, S.; Chang, Q.; Wang, D. Bringing Machine Learning to Research on Intellectual and Developmental Disabilities: Taking Inspiration from Neurological Diseases. J. Neurodev. Disord. 2022, 14, 28. [Google Scholar] [CrossRef]
- Gemein, L.A.W.; Schirrmeister, R.T.; Chrabąszcz, P.; Wilson, D.; Boedecker, J.; Schulze-Bonhage, A.; Hutter, F.; Ball, T. Machine-Learning-Based Diagnostics of EEG Pathology. NeuroImage 2020, 220, 117021. [Google Scholar] [CrossRef]
- Dev, A.; Roy, N.; Islam, K.; Biswas, C.; Ahmed, H.U.; Amin, A.; Sarker, F.; Vaidyanathan, R.; Mamun, K.A. Exploration of EEG-Based Depression Biomarkers Identification Techniques and Their Applications: A Systematic Review. IEEE Access 2022, 10, 16756–16781. [Google Scholar] [CrossRef]
- Noor, N.S.E.M.; Ibrahim, H. Machine Learning Algorithms and Quantitative Electroencephalography Predictors for Outcome Prediction in Traumatic Brain Injury: A Systematic Review. IEEE Access 2020, 8, 102075–102092. [Google Scholar] [CrossRef]
- Saeidi, M.; Karwowski, W.; Farahani, F.V.; Fiok, K.; Taiar, R.; Hancock, P.A.; Al-Juaid, A. Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sci. 2021, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Vahid, A.; Mückschel, M.; Stober, S.; Stock, A.-K.; Beste, C. Applying Deep Learning to Single-Trial EEG Data Provides Evidence for Complementary Theories on Action Control. Commun. Biol. 2020, 3, 112. [Google Scholar] [CrossRef] [PubMed]
- Bussu, G.; Jones, E.J.H.; Charman, T.; Johnson, M.H.; Buitelaar, J.K. Prediction of Autism at 3 Years from Behavioural and Developmental Measures in High-Risk Infants: A Longitudinal Cross-Domain Classifier Analysis. J. Autism. Dev. Disord. 2018, 48, 2418–2433. [Google Scholar] [CrossRef]
- Musa, A.B. Comparative Study on Classification Performance between Support Vector Machine and Logistic Regression. Int. J. Mach. Learn. Cyber. 2013, 4, 13–24. [Google Scholar] [CrossRef]
- Molnar, C. Interpretable Machine Learning: A Guide for Making Black Box Models Explainable, 2nd ed.; 2022; ISBN 979-8411463330. [Google Scholar]
- Liao, M.; Duan, H.; Wang, G. Application of Machine Learning Techniques to Detect the Children with Autism Spectrum Disorder. J. Healthc. Eng. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Hosseini, M.-P.; Hosseini, A.; Ahi, K. A Review on Machine Learning for EEG Signal Processing in Bioengineering. IEEE Rev. Biomed. Eng. 2021, 14, 204–218. [Google Scholar] [CrossRef]
- Russell, S.J.; Norvig, P. Artificial Intelligence: A Modern Approach, 3rd ed.; Pearson: New York, NY, USA, 2009; ISBN 978-0-13-604259-4. [Google Scholar]
- Abdel Hameed, M.; Hassaballah, M.; Hosney, M.E.; Alqahtani, A. An AI-Enabled Internet of Things Based Autism Care System for Improving Cognitive Ability of Children with Autism Spectrum Disorders. Comput. Intell. Neurosci. 2022, 2022, 1–12. [Google Scholar] [CrossRef]
- Sklearn.Svm.SVC. Available online: https://scikit-learn/stable/modules/generated/sklearn.svm.SVC.html (accessed on 19 August 2022).
- Garg, A.; Mago, V. Role of Machine Learning in Medical Research: A Survey. Comput. Sci. Rev. 2021, 40, 100370. [Google Scholar] [CrossRef]
- Baygin, M.; Dogan, S.; Tuncer, T.; Datta Barua, P.; Faust, O.; Arunkumar, N.; Abdulhay, E.W.; Emma Palmer, E.; Rajendra Acharya, U. Automated ASD Detection Using Hybrid Deep Lightweight Features Extracted from EEG Signals. Comput. Biol. Med. 2021, 134, 104548. [Google Scholar] [CrossRef]
- Catarino, A.; Andrade, A.; Churches, O.; Wagner, A.P.; Baron-Cohen, S.; Ring, H. Task-Related Functional Connectivity in Autism Spectrum Conditions: An EEG Study Using Wavelet Transform Coherence. Mol. Autism. 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Britton, J.; Frey, L.; Hopp, J.; Korb, P.; Koubeissi, M.; Lievens, W.; Pestana-Knight, E.; Louis, E.K.S. Electroencephalography (EEG): An Introductory Text and Atlas of Normal and Abnormal Findings in Adults, Children, and Infants; St. Louis, E.K., Frey, L., Eds.; American Epilepsy Society: Chicago, IL, USA, 2016; ISBN 978-0-9979756-0-4. [Google Scholar]
- Alhassan, S.; Soudani, A. Energy-Aware EEG-Based Scheme for Early-Age Autism Detection. In Proceedings of the 2022 2nd International Conference of Smart Systems and Emerging Technologies (SMARTTECH), Riyadh, Saudi Arabia, 9–11 May 2022; IEEE: Piscataway, NJ, USA; pp. 97–102. [Google Scholar]
- Muthukumaraswamy, S.D.; Singh, K.D. Visual Gamma Oscillations: The Effects of Stimulus Type, Visual Field Coverage and Stimulus Motion on MEG and EEG Recordings. NeuroImage 2013, 69, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.L.; Jahmunah, V.; Arunkumar, N.; Abdulhay, E.W.; Gururajan, R.; Adib, N.; Ciaccio, E.J.; Cheong, K.H.; Acharya, U.R. A Novel Automated Autism Spectrum Disorder Detection System. Complex Intell. Syst. 2021, 7, 2399–2413. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Bhat, S.; Koh, J.E.W. Application of Entropies for Automated Diagnosis of Epilepsy Using EEG Signals: A Review. Knowl. Based Syst. 2015, 88, 85–96. [Google Scholar] [CrossRef]
- Contiki-NG · GitHub. Available online: https://github.com/contiki-ng (accessed on 12 September 2022).
- Kurniawan, A. Practical Contiki-NG; Apress: Berkeley, CA, USA, 2018; ISBN 978-1-4842-3407-5. [Google Scholar]
- Amrani, G.; Adadi, A.; Berrada, M.; Souirti, Z.; Boujraf, S. EEG Signal Analysis Using Deep Learning: A Systematic Literature Review. In Proceedings of the 2021 Fifth International Conference On Intelligent Computing in Data Sciences (ICDS), Fez, Morocco, 20–22 October 2021; pp. 1–8. [Google Scholar]
- Zaidi, S.A.R.; Hayajneh, A.M.; Hafeez, M.; Ahmed, Q.Z. Unlocking Edge Intelligence Through Tiny Machine Learning (TinyML). IEEE Access 2022, 10, 100867–100877. [Google Scholar] [CrossRef]
- Shoaran, M.; Haghi, B.A.; Taghavi, M.; Farivar, M.; Emami-Neyestanak, A. Energy-Efficient Classification for Resource-Constrained Biomedical Applications. IEEE J. Emerg. Sel. Top. Circuits Syst. 2018, 8, 693–707. [Google Scholar] [CrossRef]








| Wavelet Coefficient and Its Frequency | EEG Approximate Label |
|---|---|
| D1 (25 Hz–50 Hz) | Gamma |
| D2 (12 Hz–25 Hz) | Beta |
| D3 (6 Hz–12 Hz) | Alpha |
| D4 (3 Hz–6 Hz) | Theta |
| A4 (0.1 Hz–3 Hz) | Delta |
| Frequency Sub-Band | Number of Features | Number of Features after Permutation and Mann Whitney | Number of Features after Spearman Correlation |
|---|---|---|---|
| gamma | 120 | 25 | 4 |
| beta | 120 | 56 | 10 |
| alpha | 120 | 56 | 8 |
| theta | 120 | 44 | 10 |
| delta | 120 | 22 | 4 |
| Classifier | Hyperparameter | Values |
|---|---|---|
| Support Vector Machine | Kernel | linear |
| regularization parameter, C | 0.01, 0.1, 1, 10, 100 | |
| Logistic Regression | Solver | liblinear, newton-cg, lbfgs |
| regularization parameter, C | 0.01, 0.1, 1, 10, 100 | |
| Decision Tree | Criterion | gini, entropy |
| Splitter | random, best |
| Number of Features | Sub-Band | Acc | Sen | Spec | PPV | NPV | F1 |
|---|---|---|---|---|---|---|---|
| 1 | Beta | 83.33 | 93.33 | 73.33 | 77.78 | 91.67 | 84.85 |
| 1 | Beta | 83.33 | 93.33 | 73.33 | 77.78 | 91.67 | 84.85 |
| 1 | Alpha | 83.33 | 80 | 86.67 | 85.71 | 81.25 | 82.76 |
| 1 | Alpha | 86.67 | 86.67 | 86.67 | 86.67 | 86.67 | 86.67 |
| 2 | alpha/gamma | 93.3 | 86.67 | 100 | 100 | 88.2 | 92.86 |
| Classifier | Sub-Band | Number of Features | Channel, Features Resulting from RFE | Hyperparameter | Hyperparameter Value |
|---|---|---|---|---|---|
| SVM | Gamma | 3 | TP7, absolute Welch | Kernel C | Linear 10 |
| P8, ApEn | |||||
| F7, ApEn Normalized | |||||
| Logistic Regression | Gamma | 3 | TP7, absolute Welch | Solver C | liblinear, newton-cg, lbfgs 10, 100 |
| P8, ApEn | |||||
| F7, ApEn Normalized | |||||
| Decision Tree | Alpha | 4 | P7, Variance | criterion splitter | entropy random |
| T8, absolute Welch | |||||
| TP7, relative Welch | |||||
| TP8, ApEn Normalized |
| Study | Classifier | Acc | Sen | Spec | PPV | NPV | F1 |
|---|---|---|---|---|---|---|---|
| Our Scheme | Threshold | 93.3 | 86.67 | 100 | 100 | 88.2 | 92.86 |
| SVM | 96.67 | 100 | 95 | 93.33 | 100 | 96.55 | |
| Logistic Regression | 96.67 | 100 | 95 | 93.33 | 100 | 96.55 | |
| Decision Tree | 96.67 | 100 | 96 | 90 | 100 | 94.74 | |
| Bosl et al. [7] | SVM | 63.33 | 95 | 35 | 59 | 90 | 72.79 |
| Gabard-Durnam et al. [22] | Logistic Regression | 73.33 | 73.33 | 75 | 58.33 | 80 | 64.98 |
| Zhao et al. [21] | SVM | 73.33 | 79.33 | 56.67 | 67.67 | 68.33 | 73.04 |
| ML Classification Algorithm | Logistic Regression/SVM | Decision Tree | |||||
|---|---|---|---|---|---|---|---|
| Sub-band, Extracted Features | gamma, absolute Welch | gamma, ApEn | gamma, ApEn Normalized | alpha, variance | alpha, absolute Welch | alpha, relative Welch | alpha, ApEn Normalized |
| CPU (ticks)1 | 322,124 | 217,377 | 217,575 | 851 | 322,096 | 322,108 | 14,957 |
| Total Time (ticks)2 | 387,115 | 294,830 | 294,830 | 98,222 | 387,115 | 387,115 | 98,222 |
| Scheme | On-Node Feature Extraction and Classification | Streaming | |
|---|---|---|---|
| Logistic Regression/SVM | Decision Tree | ||
| Total Time (ticks)1 | 976,775 | 970,674 | 43,579,790 |
| CPU (ticks)2 | 757,076 | 660,012 | 1,062,120 |
| Radio Tx (ticks)3 | 102 | 102 | 189,570 |
| Radio Rx (ticks)4 | 976,673 | 970,572 | 43,390,220 |
| Transmit Energy consumption (mJ)5 | 0.16 | 0.16 | 301.99 |
| CPU Energy consumption (mJ)6 | 693.12 | 604.26 | 972.40 |
| Total Energy consumption (mJ)7 | 2374.33 | 2274.96 | 75,957.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhassan, S.; Soudani, A.; Almusallam, M. Energy-Efficient EEG-Based Scheme for Autism Spectrum Disorder Detection Using Wearable Sensors. Sensors 2023, 23, 2228. https://doi.org/10.3390/s23042228
Alhassan S, Soudani A, Almusallam M. Energy-Efficient EEG-Based Scheme for Autism Spectrum Disorder Detection Using Wearable Sensors. Sensors. 2023; 23(4):2228. https://doi.org/10.3390/s23042228
Chicago/Turabian StyleAlhassan, Sarah, Adel Soudani, and Manan Almusallam. 2023. "Energy-Efficient EEG-Based Scheme for Autism Spectrum Disorder Detection Using Wearable Sensors" Sensors 23, no. 4: 2228. https://doi.org/10.3390/s23042228
APA StyleAlhassan, S., Soudani, A., & Almusallam, M. (2023). Energy-Efficient EEG-Based Scheme for Autism Spectrum Disorder Detection Using Wearable Sensors. Sensors, 23(4), 2228. https://doi.org/10.3390/s23042228





