A Deep Learning Image System for Classifying High Oleic Sunflower Seed Varieties
Abstract
1. Introduction
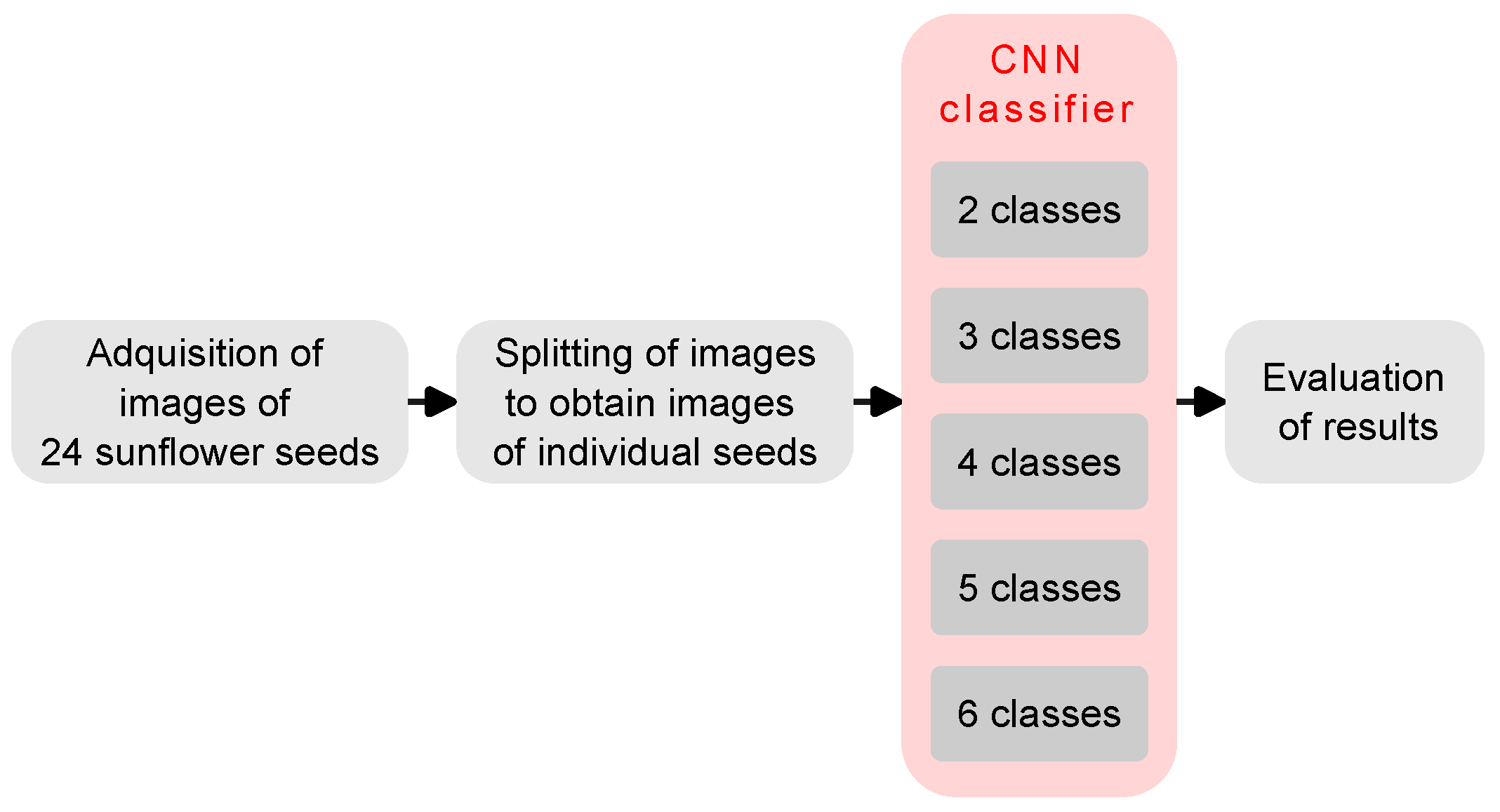
2. Materials and Methods
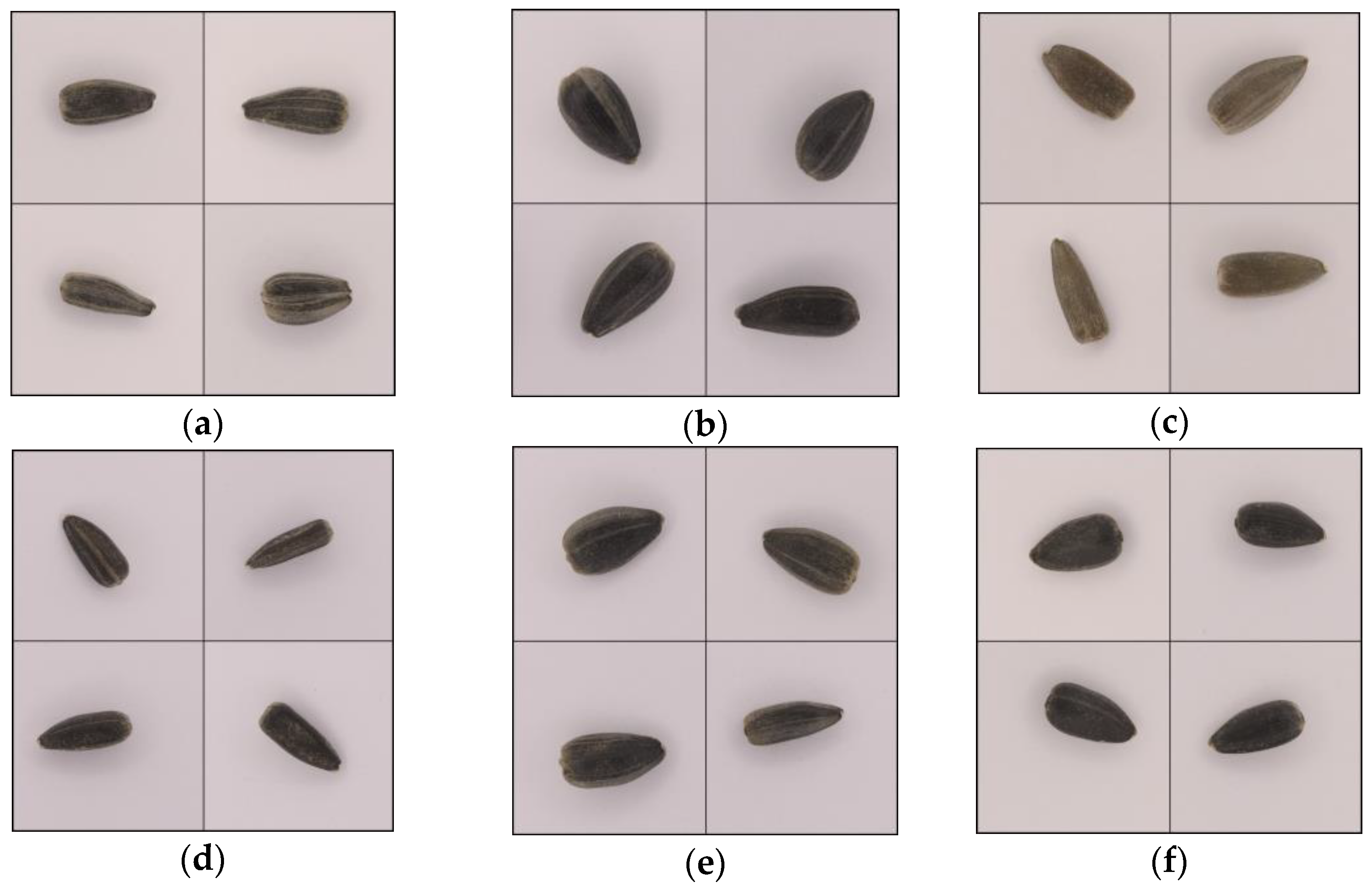
2.1. Sunflower Seeds
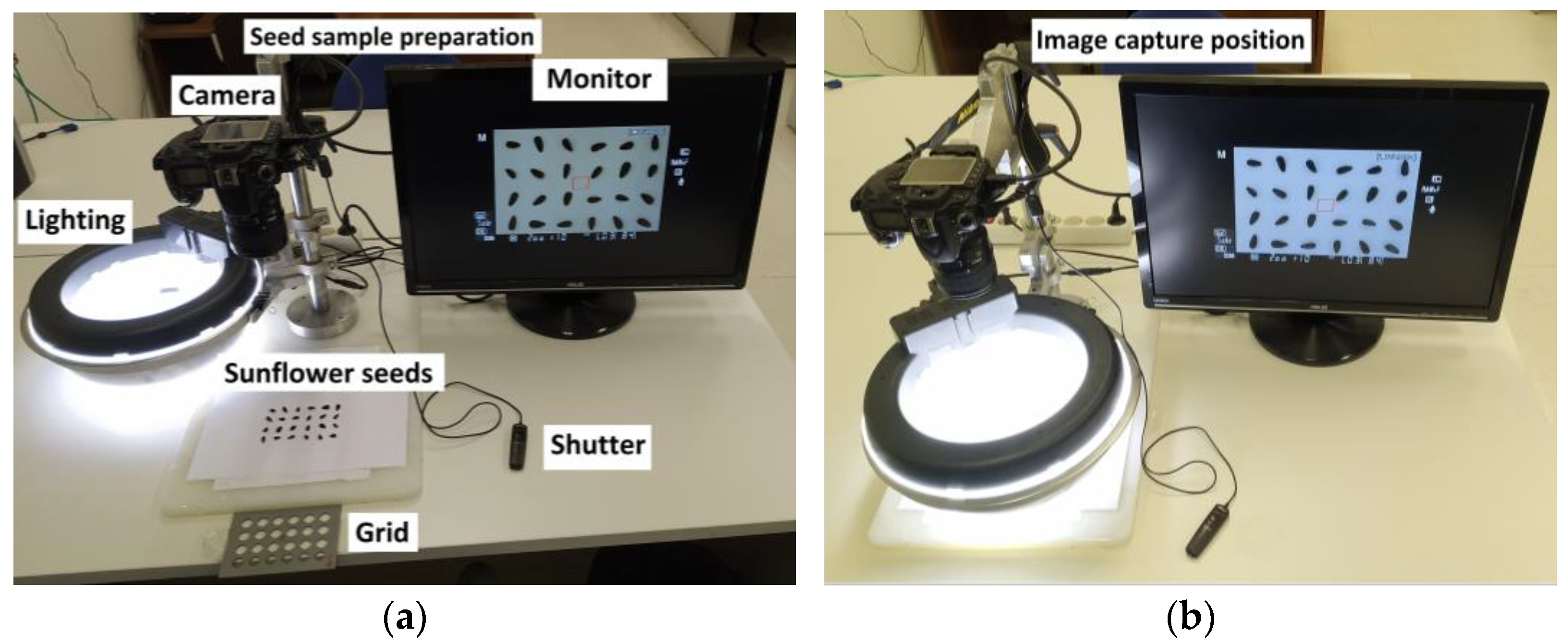
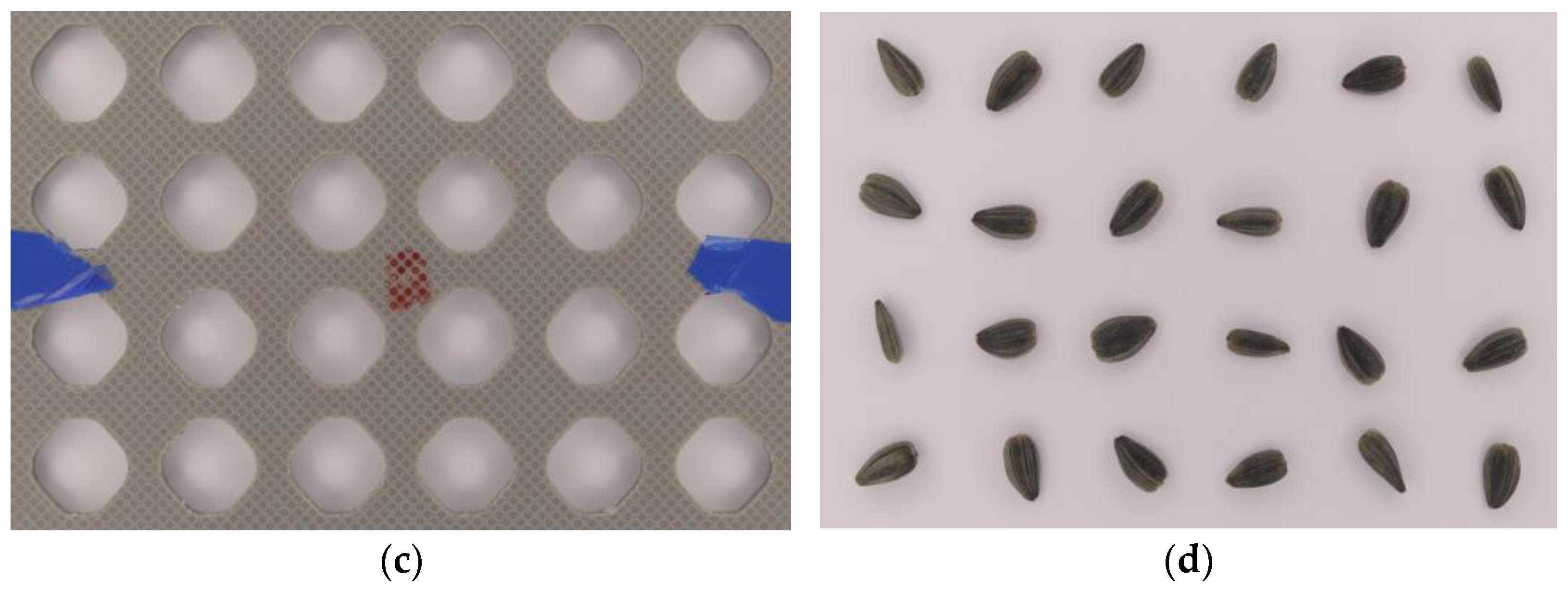
2.2. Image Acquisition Setup
2.3. Splitting of Seed Images
2.4. CNN Classifier
2.5. Evaluation of Results
2.6. Hardware Equipment and Software
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chávez-Dulanto, P.N.; Thiry, A.A.A.; Glorio-Paulet, P.; Vögler, O.; Carvalho, F.P. Increasing the Impact of Science and Technology to Provide More People with Healthier and Safer Food. Food Energy Secur. 2021, 10, 1–31. [Google Scholar] [CrossRef]
- Vollset, S.E.; Goren, E.; Yuan, C.W.; Cao, J.; Smith, A.E.; Hsiao, T.; Bisignano, C.; Azhar, G.S.; Castro, E.; Chalek, J.; et al. Fertility, Mortality, Migration, and Population Scenarios for 195 Countries and Territories from 2017 to 2100: A Forecasting Analysis for the Global Burden of Disease Study. Lancet 2020, 396, 1285–1306. [Google Scholar] [CrossRef]
- USDA Oilseeds: World Markets and Trade. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/tx31qh68h/k06999499/gm80jz36g/oilseeds.pdf (accessed on 8 February 2022).
- Zeb, A. A Comprehensive Review on Different Classes of Polyphenolic Compounds Present in Edible Oils. Food Res. Int. 2021, 143, 110312. [Google Scholar] [CrossRef]
- Gupta, M.K. Sunflower Oil: History, Applications and Trends. Lipid Technol. 2014, 26, 260–263. [Google Scholar] [CrossRef]
- Bowen, K.J.; Kris-Etherton, P.M.; West, S.G.; Fleming, J.A.; Connelly, P.W.; Lamarche, B.; Couture, P.; Jenkins, D.J.A.; Taylor, C.G.; Zahradka, P.; et al. Diets Enriched with Conventional or High-Oleic Acid Canola Oils Lower Atherogenic Lipids and Lipoproteins Compared to a Diet with a Western Fatty Acid Profile in Adults with Central Adiposity. J. Nutr. 2019, 149, 471–478. [Google Scholar] [CrossRef]
- Raatz, S.K.; Conrad, Z.; Jahns, L.; Belury, M.A.; Picklo, M.J. Modeled Replacement of Traditional Soybean and Canola Oil with High-Oleic Varieties Increases Monounsaturated Fatty Acid and Reduces Both Saturated Fatty Acid and Polyunsaturated Fatty Acid Intake in the US Adult Population. Am. J. Clin. Nutr. 2018, 108, 594–602. [Google Scholar] [CrossRef]
- Downs, S.M.; Bloem, M.Z.; Zheng, M.; Catterall, E.; Thomas, B.; Veerman, L.; Wu, J.H.Y. The Impact of Policies to Reduce Trans Fat Consumption: A Systematic Review of the Evidence. Curr. Dev. Nutr. 2017, 1, 1–10. [Google Scholar] [CrossRef]
- Zambelli, A. Current Status of High Oleic Seed Oils in Food Processing. JAOCS 2021, 98, 129–137. [Google Scholar] [CrossRef]
- Petraru, A.; Ursachi, F.; Amariei, S. Nutritional Characteristics Assessment of Sunflower Seeds, Oil and Cake. Perspective of Using Sunflower Oilcakes as a Functional Ingredient. Plants 2021, 10, 2487. [Google Scholar] [CrossRef]
- Arrutia, F.; Binner, E.; Williams, P.; Waldron, K.W. Oilseeds beyond Oil: Press Cakes and Meals Supplying Global Protein Requirements. Trends Food Sci. Technol. 2020, 100, 88–102. [Google Scholar] [CrossRef]
- De Morais Oliveira, V.R.; de Arruda, A.M.V.; Silva, L.N.S.; de Souza, J.B.F.; de Queiroz, J.P.A.F.; da Silva Melo, A.; Holanda, J.S. Sunflower Meal as a Nutritional and Economically Viable Substitute for Soybean Meal in Diets for Free-Range Laying Hens. Anim. Feed. Sci. Technol. 2016, 220, 103–108. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. Oilseed Crop Sunflower (Helianthus Annuus) as a Source of Food: Nutritional and Health Benefits. Food Sci. Nutr. 2020, 8, 4666–4684. [Google Scholar] [CrossRef] [PubMed]
- Anastopoulos, I.; Ighalo, J.O.; Adaobi Igwegbe, C.; Giannakoudakis, D.A.; Triantafyllidis, K.S.; Pashalidis, I.; Kalderis, D. Sunflower-Biomass Derived Adsorbents for Toxic/Heavy Metals Removal from (Waste) Water. J. Mol. Liq. 2021, 342, 117540. [Google Scholar] [CrossRef]
- Elkelawy, M.; Bastawissi, H.A.E.; Esmaeil, K.K.; Radwan, A.M.; Panchal, H.; Sadasivuni, K.K.; Suresh, M.; Israr, M. Maximization of Biodiesel Production from Sunflower and Soybean Oils and Prediction of Diesel Engine Performance and Emission Characteristics through Response Surface Methodology. Fuel 2020, 266, 117072. [Google Scholar] [CrossRef]
- Jadhav, J.V.; Anbu, P.; Yadav, S.; Pratap, A.P.; Kale, S.B. Sunflower Acid Oil-Based Production of Rhamnolipid Using Pseudomonas Aeruginosa and Its Application in Liquid Detergents. J. Surfactants Deterg. 2019, 22, 463–476. [Google Scholar] [CrossRef]
- Rauf, S.; Jamil, N.; Tariq, S.A.; Khan, M.; Kausar, M.; Kaya, Y. Progress in Modification of Sunflower Oil to Expand Its Industrial Value. J. Sci. Food Agric. 2017, 97, 1997–2006. [Google Scholar] [CrossRef]
- Alberio, C.; Izquierdo, N.G.; Galella, T.; Zuil, S.; Reid, R.; Zambelli, A.; Aguirrezábal, L.A.N. A New Sunflower High Oleic Mutation Confers Stable Oil Grain Fatty Acid Composition across Environments. Eur. J. Agron. 2016, 73, 25–33. [Google Scholar] [CrossRef]
- Attia, Z.; Pogoda, C.S.; Reinert, S.; Kane, N.C.; Hulke, B.S. Breeding for Sustainable Oilseed Crop Yield and Quality in a Changing Climate. Theor. Appl. Genet. 2021, 134, 1817–1827. [Google Scholar] [CrossRef]
- Diovisalvi, N.; Calvo, N.R.; Izquierdo, N.; Echeverría, H.; Divito, G.A.; García, F. Effects of Genotype and Nitrogen Availability on Grain Yield and Quality in Sunflower. Agron. J. 2018, 110, 1532–1543. [Google Scholar] [CrossRef]
- Krig, S. Computer Vision Metrics: Survey, Taxonomy, and Analysis; Springer: Berlin/Heidelberg, Germany, 2014; Volume 9781430259, ISBN 9781430259305. [Google Scholar]
- Velesaca, H.O.; Suárez, P.L.; Mira, R.; Sappa, A.D. Computer Vision Based Food Grain Classification: A Comprehensive Survey. Comput. Electron. Agric. 2021, 187, 106287. [Google Scholar] [CrossRef]
- Sabanci, K.; Kayabasi, A.; Toktas, A. Computer Vision-Based Method for Classification of Wheat Grains Using Artificial Neural Network. J. Sci. Food Agric. 2017, 97, 2588–2593. [Google Scholar] [CrossRef]
- Koklu, M.; Ozkan, I.A. Multiclass Classification of Dry Beans Using Computer Vision and Machine Learning Techniques. Comput. Electron. Agric. 2020, 174, 105507. [Google Scholar] [CrossRef]
- Khatri, A.; Agrawal, S.; Chatterjee, J.M. Wheat Seed Classification: Utilizing Ensemble Machine Learning Approach. Sci. Program. 2022, 2022, 1–9. [Google Scholar] [CrossRef]
- Shrestha, B.L.; Kang, Y.M.; Yu, D.; Baik, O.D. A Two-Camera Machine Vision Approach to Separating and Identifying Laboratory Sprouted Wheat Kernels. Biosyst. Eng. 2016, 147, 265–273. [Google Scholar] [CrossRef]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A Convolution Neural Network-Based Seed Classification System. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep Learning for Visual Understanding: A Review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Altuntaş, Y.; Cömert, Z.; Kocamaz, A.F. Identification of Haploid and Diploid Maize Seeds Using Convolutional Neural Networks and a Transfer Learning Approach. Comput. Electron. Agric. 2019, 163, 104874. [Google Scholar] [CrossRef]
- Kozłowski, M.; Górecki, P.; Szczypiński, P.M. Varietal Classification of Barley by Convolutional Neural Networks. Biosyst. Eng. 2019, 184, 155–165. [Google Scholar] [CrossRef]
- Laabassi, K.; Belarbi, M.A.; Mahmoudi, S.; Mahmoudi, S.A.; Ferhat, K. Wheat Varieties Identification Based on a Deep Learning Approach. J. Saudi Soc. Agric. Sci. 2021, 20, 281–289. [Google Scholar] [CrossRef]
- Eryigit, R.; Tugrul, B. Performance of Various Deep-Learning Networks in the Seed. Symmetry 2021, 13, 1892. [Google Scholar] [CrossRef]
- Xu, P.; Tan, Q.; Zhang, Y.; Zha, X.; Yang, S.; Yang, R. Research on Maize Seed Classification and Recognition Based on Machine Vision and Deep Learning. Agriculture 2022, 12, 232. [Google Scholar] [CrossRef]
- Taheri-Garavand, A.; Nasiri, A.; Fanourakis, D.; Fatahi, S.; Omid, M.; Nikoloudakis, N. Automated in Situ Seed Variety Identification via Deep Learning: A Case Study in Chickpea. Plants 2021, 10, 1406. [Google Scholar] [CrossRef]
- Zhao, G.; Quan, L.; Li, H.; Feng, H.; Li, S.; Zhang, S.; Liu, R. Real-Time Recognition System of Soybean Seed Full-Surface Defects Based on Deep Learning. Comput. Electron. Agric. 2021, 187, 106230. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety Identification of Single Rice Seed Using Hyperspectral Imaging Combined with Convolutional Neural Network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Nie, P.; Zhang, J.; Feng, X.; Yu, C.; He, Y. Classification of Hybrid Seeds Using Near-Infrared Hyperspectral Imaging Technology Combined with Deep Learning. Sens. Actuators B Chem. 2019, 296, 126630. [Google Scholar] [CrossRef]
- Loddo, A.; Loddo, M.; di Ruberto, C. A Novel Deep Learning Based Approach for Seed Image Classification and Retrieval. Comput. Electron. Agric. 2021, 187, 106269. [Google Scholar] [CrossRef]
- Gao, J.; Liu, C.; Han, J.; Lu, Q.; Wang, H.; Zhang, J.; Bai, X.; Luo, J. Identification Method of Wheat Cultivars by Using a Convolutional Neural Network Combined with Images of Multiple Growth Periods of Wheat. Symmetry 2021, 13, 2012. [Google Scholar] [CrossRef]
- Przybyło, J.; Jabłoński, M. Using Deep Convolutional Neural Network for Oak Acorn Viability Recognition Based on Color Images of Their Sections. Comput. Electron. Agric. 2019, 156, 490–499. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep Learning in Agriculture: A Survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Lin, P.; Li, X.L.; Chen, Y.M.; He, Y. A Deep Convolutional Neural Network Architecture for Boosting Image Discrimination Accuracy of Rice Species. Food Bioproc. Tech. 2018, 11, 765–773. [Google Scholar] [CrossRef]
- Koklu, M.; Cinar, I.; Taspinar, Y.S. Classification of Rice Varieties with Deep Learning Methods. Comput. Electron. Agric. 2021, 187, 106285. [Google Scholar] [CrossRef]
- Mukasa, P.; Wakholi, C.; Akbar Faqeerzada, M.; Amanah, H.Z.; Kim, H.; Joshi, R.; Suh, H.-K.; Kim, G.; Lee, H.; Kim, M.S.; et al. Nondestructive Discrimination of Seedless from Seeded Watermelon Seeds by Using Multivariate and Deep Learning Image Analysis. Comput. Electron. Agric. 2022, 194, 106799. [Google Scholar] [CrossRef]
- Kurtulmuş, F. Identification of Sunflower Seeds with Deep Convolutional Neural Networks. J. Food Meas. Charact. 2021, 15, 1024–1033. [Google Scholar] [CrossRef]
- Li, C.; Li, H.; Liu, Z.; Li, B.; Huang, Y. SeedSortNet: A Rapid and Highly Effificient Lightweight CNN Based on Visual Attention for Seed Sorting. PeerJ. Comput. Sci. 2021, 7, e639. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Loddo, A.; di Ruberto, C. On the Efficacy of Handcrafted and Deep Features for Seed Image Classification. J. Imaging 2021, 7, 171. [Google Scholar] [CrossRef]
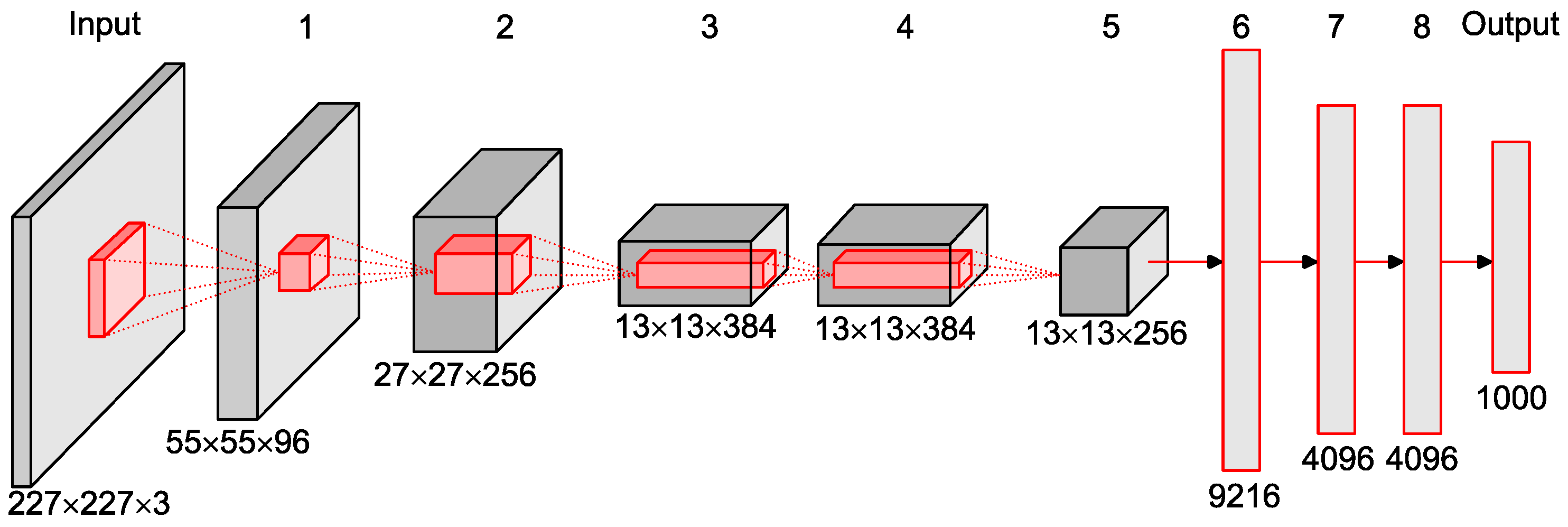


| Layer | Feature Map | Size | Kernel Size | Stride | Activation | |
|---|---|---|---|---|---|---|
| Input | Image | 1 | 227 × 227 × 3 | - | - | - |
| 1 | Convolution | 96 | 55 × 55 × 96 | 11 × 11 | 4 | relu |
| Max pooling | 96 | 27 × 27 × 96 | 3 × 3 | 2 | relu | |
| 2 | Convolution | 256 | 27 × 27 × 256 | 5 × 5 | 1 | relu |
| Max pooling | 256 | 13 × 13 × 256 | 3 × 3 | 2 | relu | |
| 3 | Convolution | 384 | 13 × 13 × 384 | 3 × 3 | 1 | relu |
| 4 | Convolution | 384 | 13 × 13 × 384 | 3 × 3 | 1 | relu |
| 5 | Convolution | 256 | 13 × 13 × 256 | 3 × 3 | 1 | relu |
| Max pooling | 256 | 6 × 6 × 256 | 3 × 3 | 2 | relu | |
| 6 | FC | - | 9216 | - | - | relu |
| 7 | FC | - | 4096 | - | - | |
| 8 | FC | - | 4096 | - | - | relu |
| Output | FC | - | 1000 | - | - | softmax |
| Factor | Value |
|---|---|
| Image input size | 227 × 227 × 3 |
| Epochs | 50 |
| Optimizer | Adam |
| Loss function | Cross-entropy |
| Batch size | 32 |
| Validation size | 30 |
| Learning rate | 0.00001 |
| Dropout | 0.5 |
| Classes | Varieties | Recall | Precision | F1-Score | AUC | Accuracy | Loss |
|---|---|---|---|---|---|---|---|
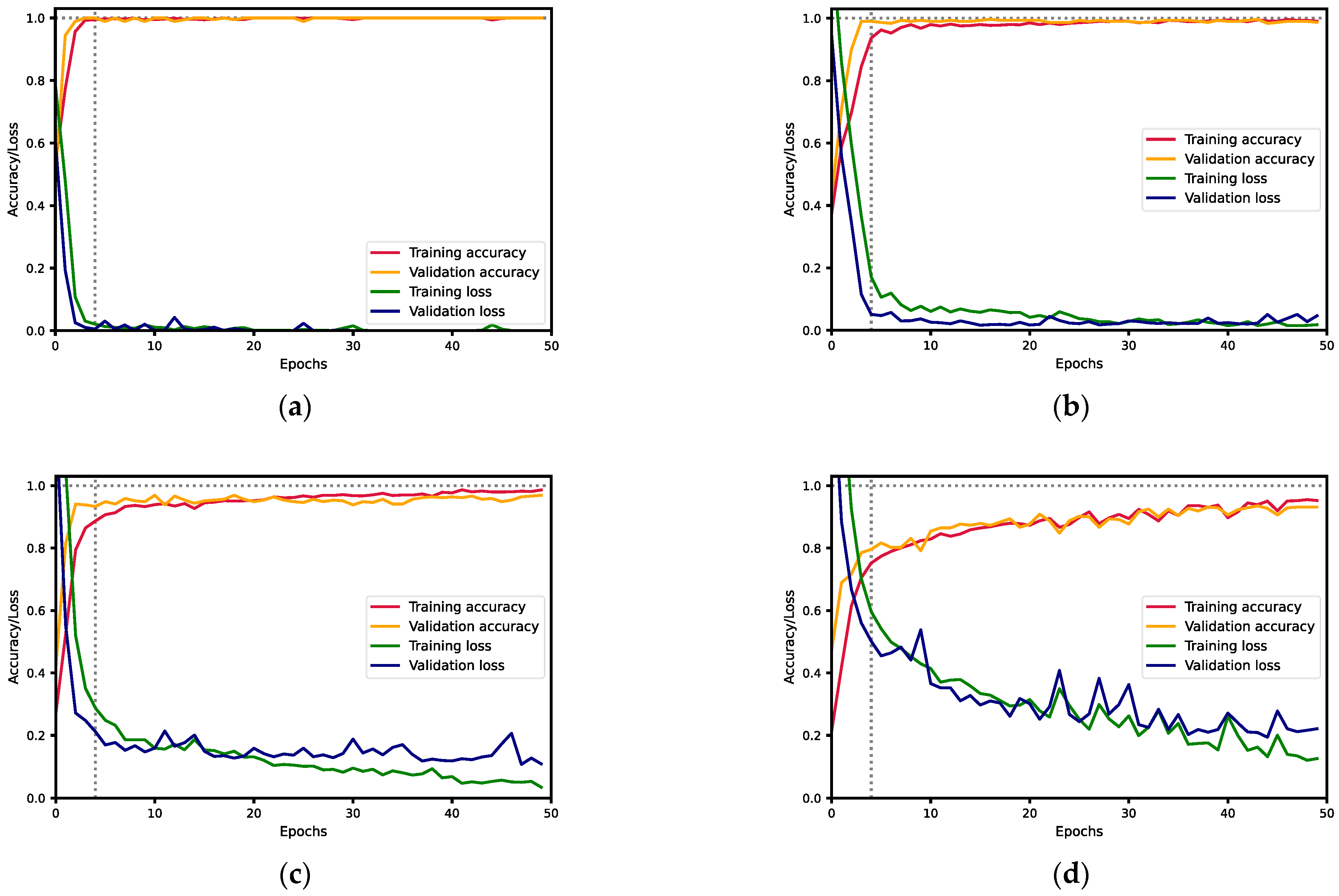
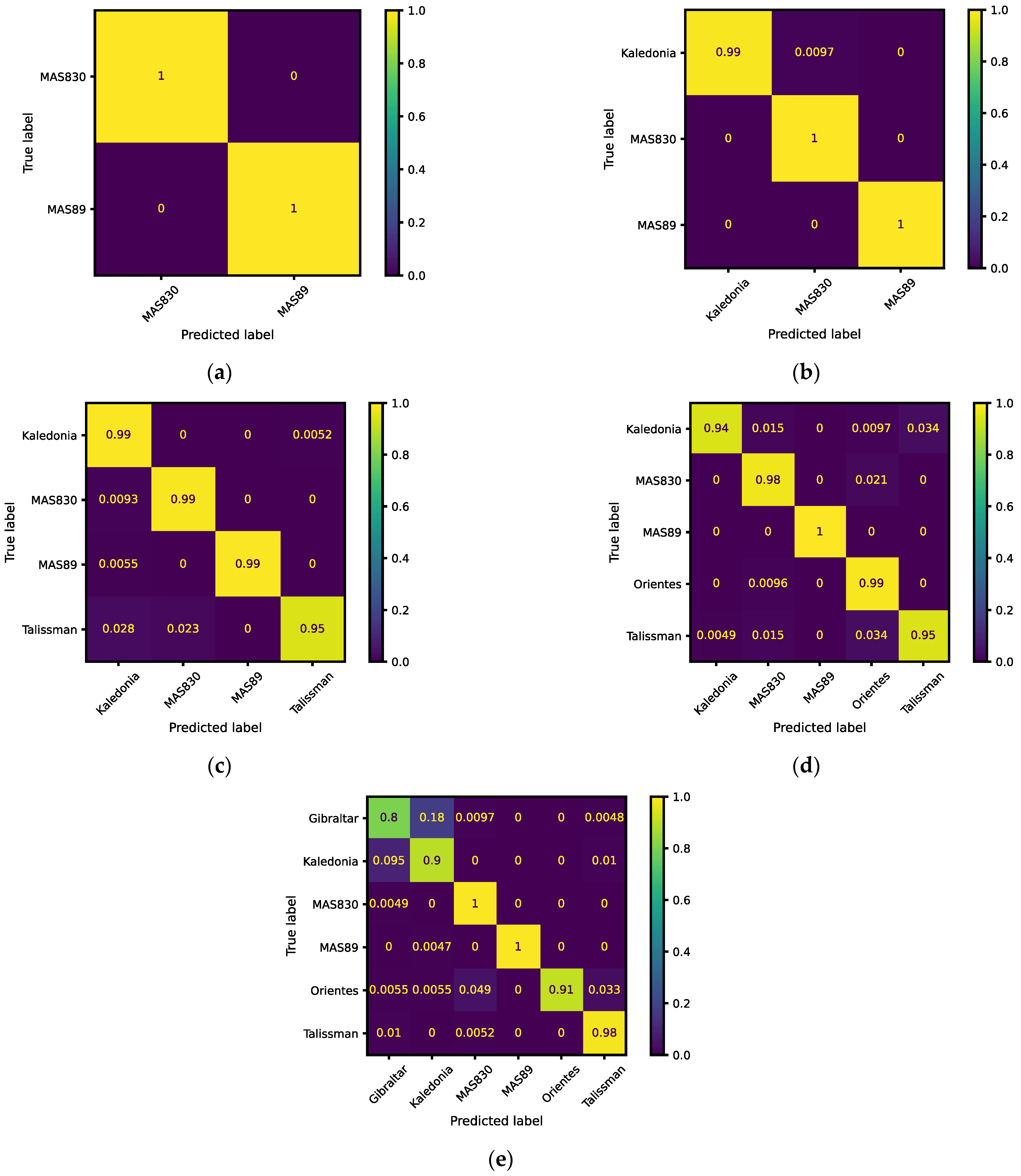
| 2 | MAS830 | 1 | 1 | 1 | 1 | 1 | 0.00005 |
| MAS89 | 1 | 1 | 1 | 1 | |||
| 3 | Kaledonia | 0.9903 | 1 | 0.9951 | 0.9952 | 0.992 | 0.02415 |
| MAS830 | 1 | 0.9896 | 0.9948 | 0.9976 | |||
| MAS89 | 1 | 1 | 1 | 1 | |||
| 4 | Kaledonia | 0.9948 | 0.9548 | 0.9744 | 0.9900 | 0.972 | 0.07590 |
| MAS830 | 0.9907 | 0.9770 | 0.9838 | 0.9911 | |||
| MAS89 | 0.9945 | 1 | 0.9972 | 0.9972 | |||
| Talissman | 0.9486 | 0.9951 | 0.9713 | 0.9734 | |||
| 5 | Kaledonia | 0.9417 | 0.9949 | 0.9676 | 0.9702 | 0.940 | 0.18497 |
| MAS830 | 0.9786 | 0.9581 | 0.9683 | 0.9844 | |||
| MAS89 | 1 | 1 | 1 | 1 | |||
| Orientes | 0.9904 | 0.9406 | 0.9649 | 0.9870 | |||
| Talissman | 0.9466 | 0.9653 | 0.9559 | 0.9689 | |||
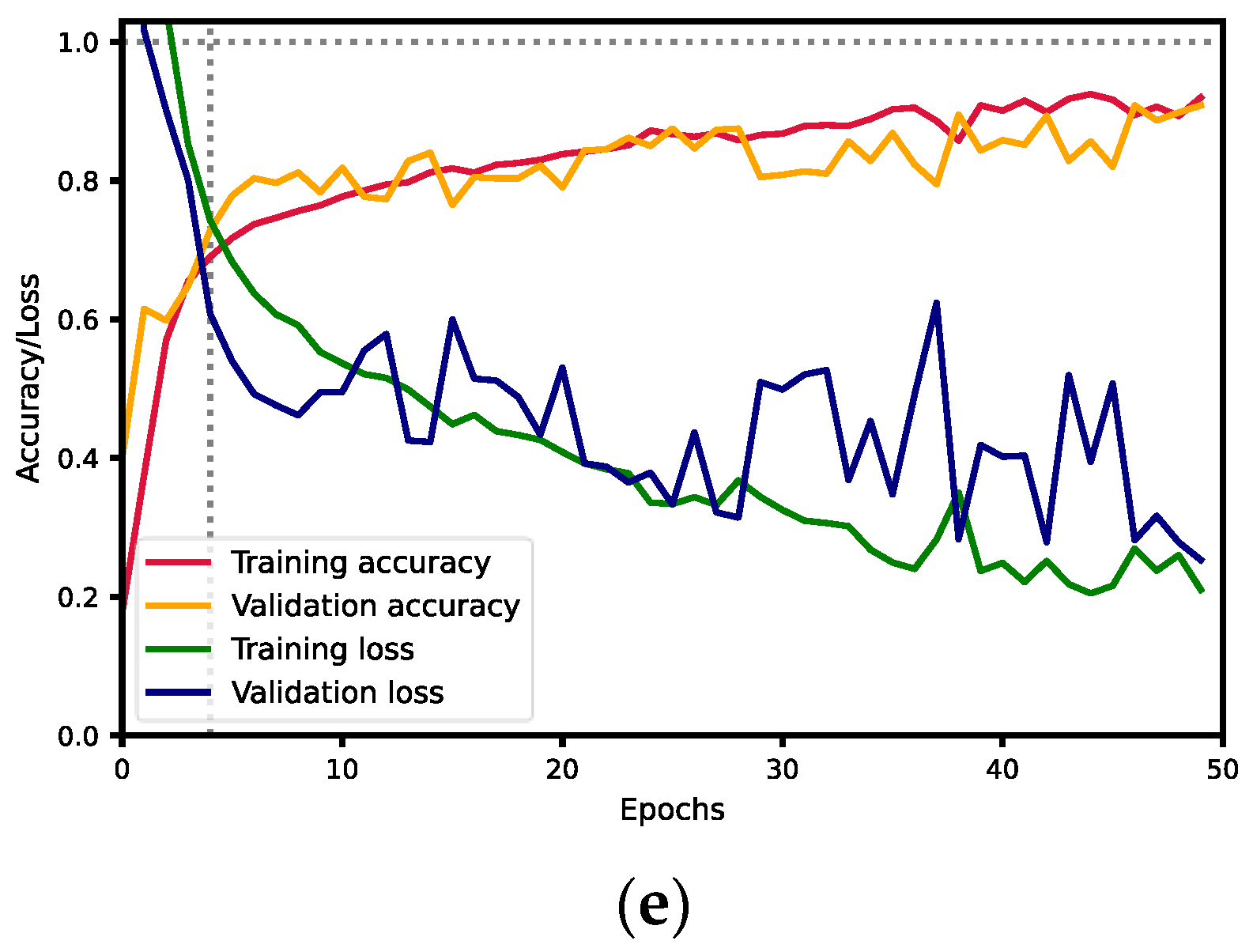
| 6 | Gibraltar | 0.8019 | 0.8783 | 0.8384 | 0.8894 | 0.895 | 0.29350 |
| Kaledonia | 0.8955 | 0.8182 | 0.8551 | 0.9277 | |||
| MAS830 | 0.9951 | 0.9444 | 0.9691 | 0.9915 | |||
| MAS89 | 0.9953 | 1 | 0.9977 | 0.9977 | |||
| Orientes | 0.9066 | 1 | 0.9510 | 0.9533 | |||
| Talissman | 0.9843 | 0.9543 | 0.9691 | 0.9877 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrio-Conde, M.; Zanella, M.A.; Aguiar-Perez, J.M.; Ruiz-Gonzalez, R.; Gomez-Gil, J. A Deep Learning Image System for Classifying High Oleic Sunflower Seed Varieties. Sensors 2023, 23, 2471. https://doi.org/10.3390/s23052471
Barrio-Conde M, Zanella MA, Aguiar-Perez JM, Ruiz-Gonzalez R, Gomez-Gil J. A Deep Learning Image System for Classifying High Oleic Sunflower Seed Varieties. Sensors. 2023; 23(5):2471. https://doi.org/10.3390/s23052471
Chicago/Turabian StyleBarrio-Conde, Mikel, Marco Antonio Zanella, Javier Manuel Aguiar-Perez, Ruben Ruiz-Gonzalez, and Jaime Gomez-Gil. 2023. "A Deep Learning Image System for Classifying High Oleic Sunflower Seed Varieties" Sensors 23, no. 5: 2471. https://doi.org/10.3390/s23052471
APA StyleBarrio-Conde, M., Zanella, M. A., Aguiar-Perez, J. M., Ruiz-Gonzalez, R., & Gomez-Gil, J. (2023). A Deep Learning Image System for Classifying High Oleic Sunflower Seed Varieties. Sensors, 23(5), 2471. https://doi.org/10.3390/s23052471









