An Optimized Active Sampling Procedure for Aerobiological DNA Studies
Abstract
:1. Introduction
2. Methodology
2.1. Bioaerosol Sampler
2.2. Commercial Filter Selection
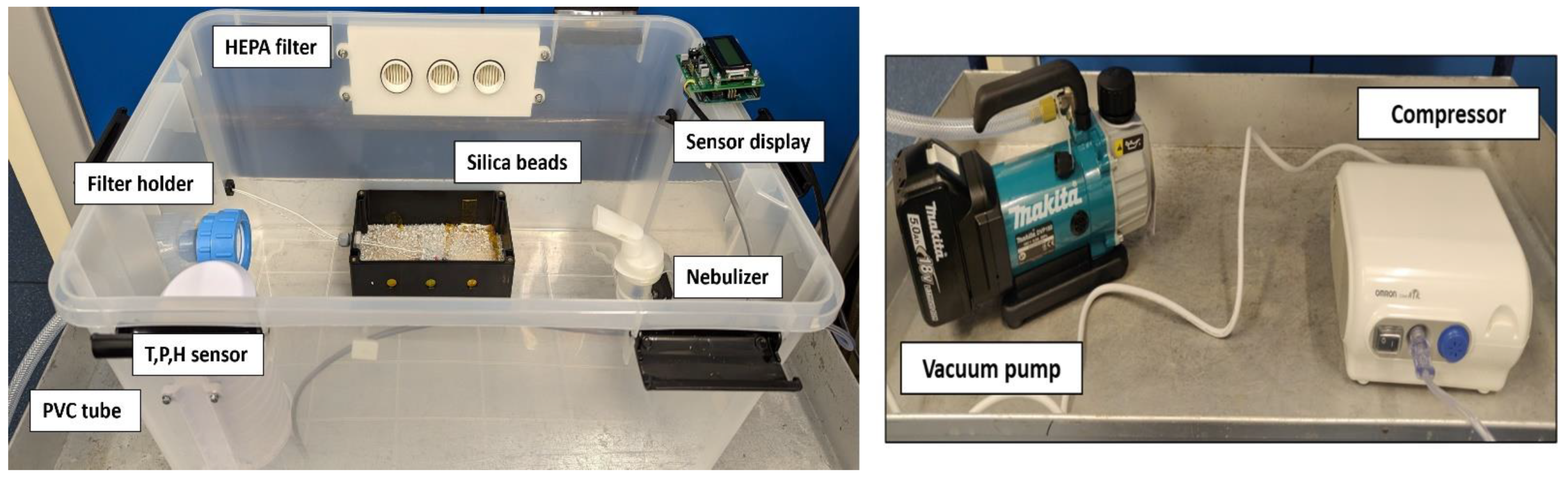
2.3. Bioaerosol Chamber
2.4. Bioaerosol Supernatant and Controls
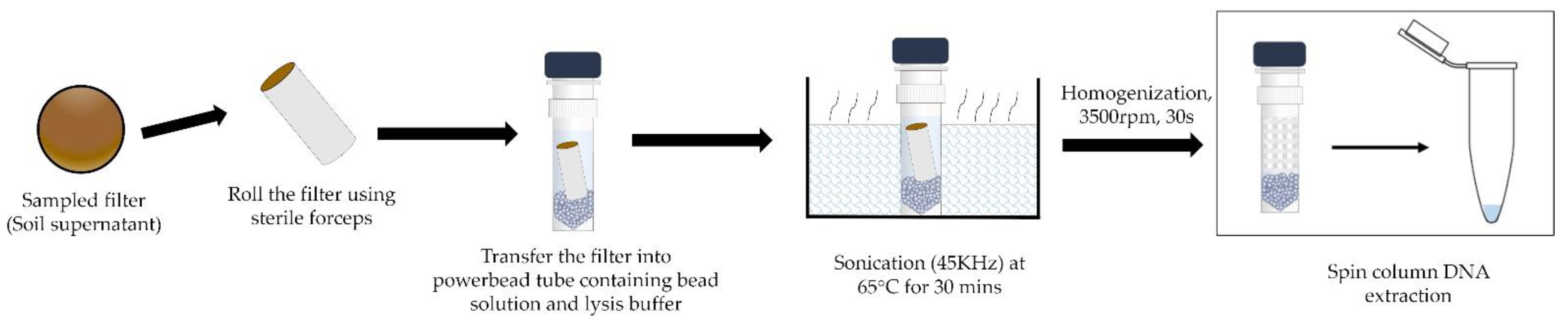
2.5. DNA Extraction
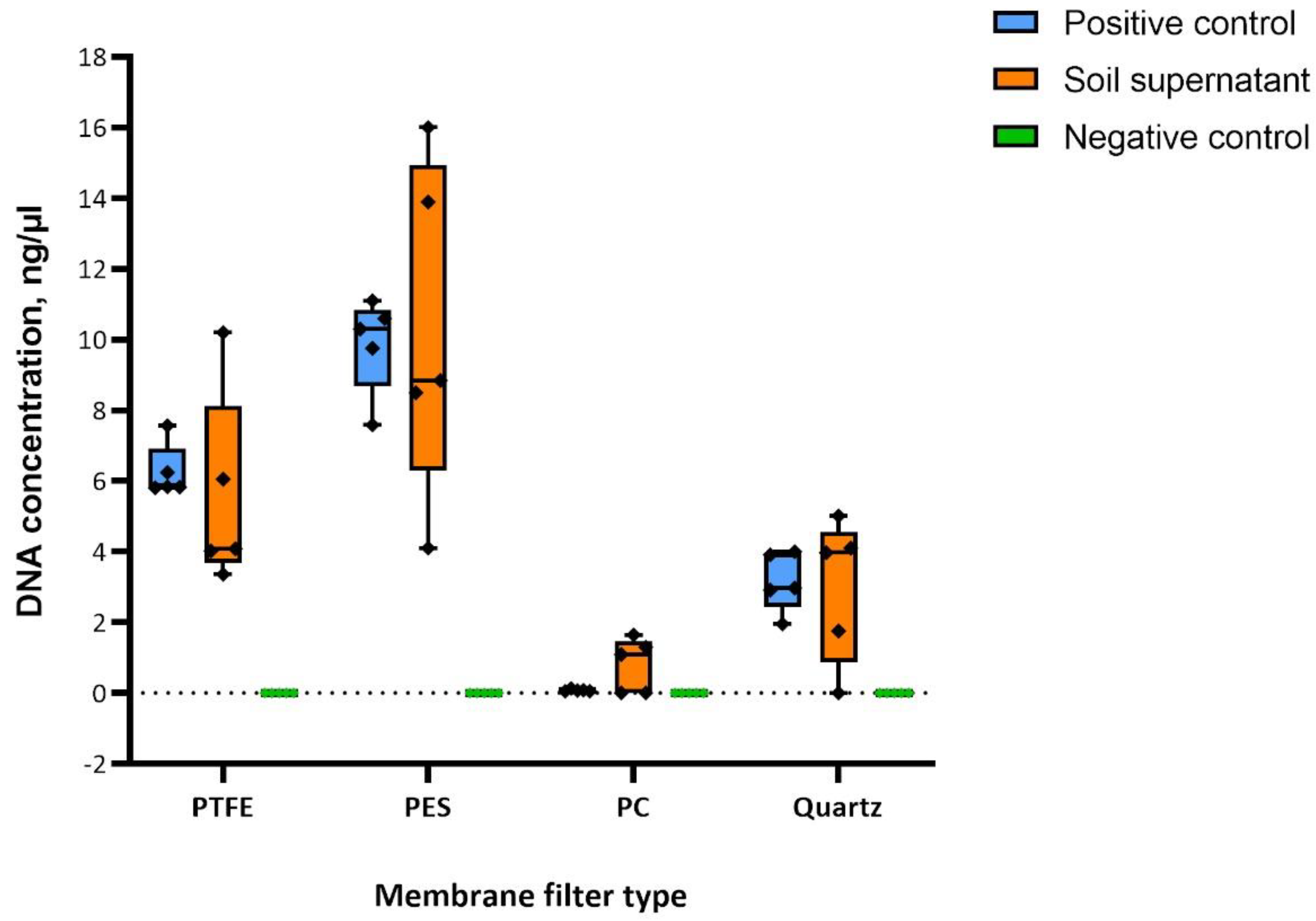
3. Results
3.1. Bioaerosol Chamber
3.2. Outdoor Environment
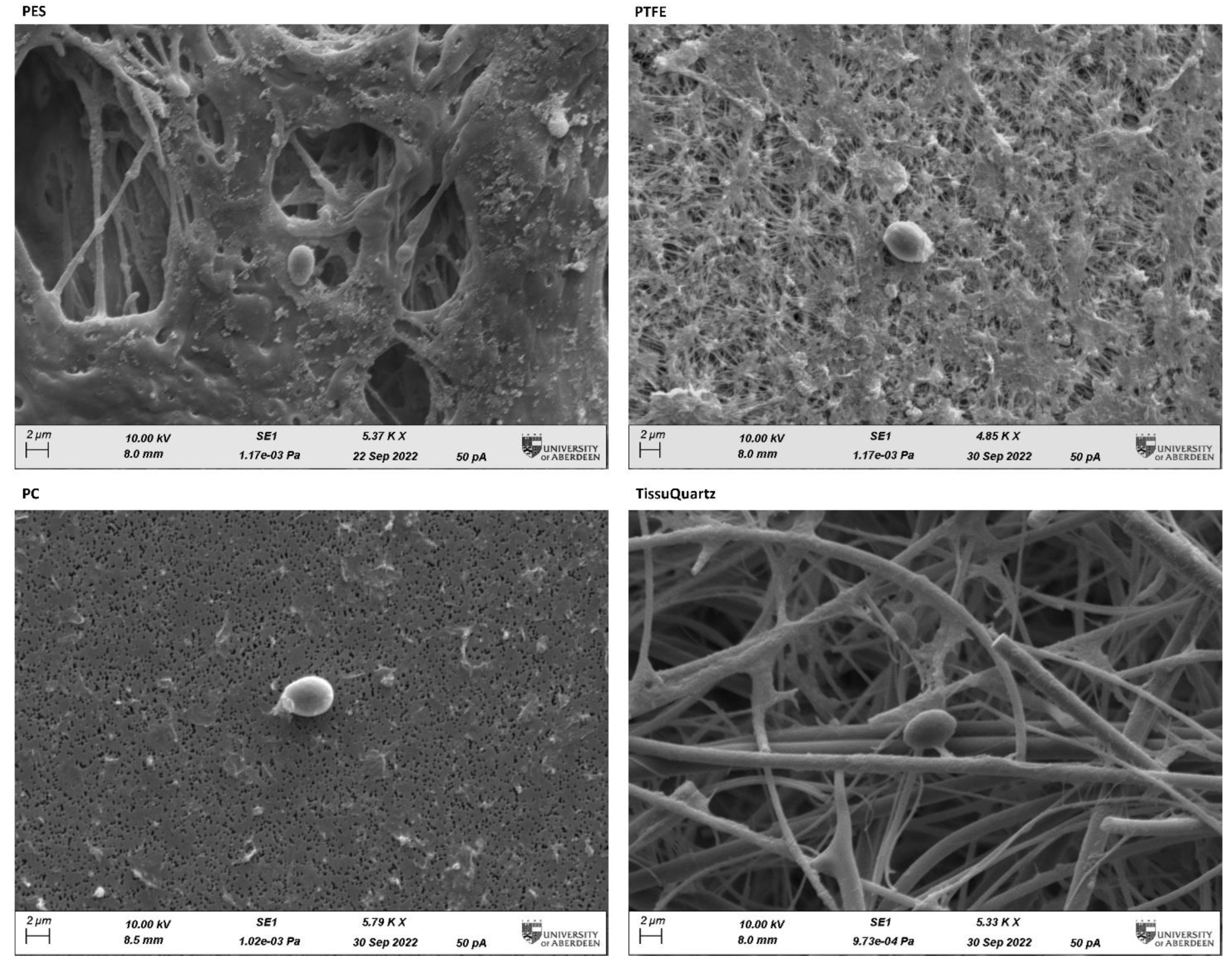
3.3. Post-Sampling Filter Micro-Scale Characterization
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Douwes, J.; Eduard, W.; Thorne, P.S. Bioaerosols. Int. Encycl. Public Health 2008, 287–297. [Google Scholar] [CrossRef]
- Yao, M. Bioaerosol: A Bridge and Opportunity for Many Scientific Research Fields. J. Aerosol Sci. 2018, 115, 108–112. [Google Scholar] [CrossRef]
- Ferguson, R.M.W.; Garcia-Alcega, S.; Coulon, F.; Dumbrell, A.J.; Whitby, C.; Colbeck, I. Bioaerosol Biomonitoring: Sampling Optimization for Molecular Microbial Ecology. Mol. Ecol. Resour. 2019, 19, 672–690. [Google Scholar] [CrossRef]
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the Earth System: Climate, Health, and Ecosystem Interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.D.; Lednicky, J.A.; Torremorell, M.; Gray, G.C. The Use of Bioaerosol Sampling for Airborne Virus Surveillance in Swine Production Facilities: A Mini Review. Front. Vet. Sci. 2017, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, J.; Gallegos-Graves, L.V.; Gans, J.; Morse, S.A.; Pillai, S.; Anderson, K.; Hodge, D.R. Evaluation of DNA Extraction Methods to Detect Bacterial Targets in Aerosol Samples. J. Microbiol. Methods 2018, 153, 48–53. [Google Scholar] [CrossRef]
- Luhung, I.; Wu, Y.; Ng, C.K.; Miller, D.; Cao, B.; Chang, V.W.-C. Protocol Improvements for Low Concentration DNA-Based Bioaerosol Sampling and Analysis. PLoS ONE 2015, 10, e0141158. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-Y.; Kim, T.G. Comparison of Five Membrane Filters to Collect Bioaerosols for Airborne Microbiome Analysis. J. Appl. Microbiol. 2021, 131, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Miaskiewicz-Peska, E.; Lebkowska, M. Comparison of Aerosol and Bioaerosol Collection on Air Filters. Aerobiologia 2012, 28, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lev, S.; Ruzer, N.H.H. Aerosols Handbook: Measurement, Dosimetry, and Health Effects, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012; ISBN 9781439855195. [Google Scholar]
- Pogner, C.; Konlechner, A.; Unterwurzacher, V.; Kolk, A.; Hinker, M.; Mölter, L.; Strauss, J.; Gorfer, M.; Strauss-Goller, S. A Novel Laminar-Flow-Based Bioaerosol Test System to Determine Biological Sampling Efficiencies of Bioaerosol Samplers. Aerosol Sci. Technol. 2019, 53, 355–370. [Google Scholar] [CrossRef]
- Trunov, M.; Trakumas, S.; Willeke, K.; Grinshpun, S.A.; Reponen, T. Collection of Bioaerosol Particles by Impaction: Effect of Fungal Spore Agglomeration and Bounce. Aerosol Sci. Technol. 2001, 35, 617–624. [Google Scholar] [CrossRef]
- Manibusan, S.; Mainelis, G. Passive Bioaerosol Samplers: A Complementary Tool for Bioaerosol Research. A Review. J. Aerosol Sci. 2022, 163, 105992. [Google Scholar] [CrossRef] [PubMed]
- Mainelis, G. Bioaerosol Sampling: Classical Approaches, Advances, and Perspectives. Aerosol Sci. Technol. 2020, 54, 496–519. [Google Scholar] [CrossRef]
- Morgan, J.L.; Darling, A.E.; Eisen, J.A. Metagenomic Sequencing of an In Vitro-Simulated Microbial Community. PLoS ONE 2010, 5, e10209. [Google Scholar] [CrossRef]
- Jiang, W.; Liang, P.; Wang, B.; Fang, J.; Lang, J.; Tian, G.; Jiang, J.; Zhu, T.F. Optimized DNA Extraction and Metagenomic Sequencing of Airborne Microbial Communities. Nat. Protoc. 2015, 10, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Mescioglu, E.; Paytan, A.; Mitchell, B.W.; Griffin, D.W. Efficiency of Bioaerosol Samplers: A Comparison Study. Aerobiologia 2021, 37, 447–459. [Google Scholar] [CrossRef]
- Galès, A.; Bru-Adan, V.; Godon, J.-J.; Delabre, K.; Catala, P.; Ponthieux, A.; Chevallier, M.; Birot, E.; Steyer, J.-P.; Wéry, N. Predominance of Single Bacterial Cells in Composting Bioaerosols. Atmos. Environ. 2015, 107, 225–232. [Google Scholar] [CrossRef]
- Pankhurst, L.J.; Whitby, C.; Pawlett, M.; Larcombe, L.D.; McKew, B.; Deacon, L.J.; Morgan, S.L.; Villa, R.; Drew, G.H.; Tyrrel, S.; et al. Temporal and Spatial Changes in the Microbial Bioaerosol Communities in Green-Waste Composting. FEMS Microbiol. Ecol. 2012, 79, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Soo, J.-C.; Monaghan, K.; Lee, T.; Kashon, M.; Harper, M. Air Sampling Filtration Media: Collection Efficiency for Respirable Size-Selective Sampling. Aerosol Sci. Technol. 2016, 50, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Unterwurzacher, V.; Bruck, S.; Biedermann, M.; Pogner, C.; Konlechner, A.; Tondl, G.; Berger, H.; Pfeifer, C.; Strauss, J.; Gorfer, M.; et al. Development and Validation of a Simple Bioaerosol Collection Filter System Using a Conventional Vacuum Cleaner for Sampling. Aerosol Sci. Eng. 2021, 5, 404–418. [Google Scholar] [CrossRef]
- Bøifot, K.O.; Gohli, J.; Moen, L.V.; Dybwad, M. Performance Evaluation of a New Custom, Multi-Component DNA Isolation Method Optimized for Use in Shotgun Metagenomic Sequencing-Based Aerosol Microbiome Research. Environ. Microbiome 2020, 15, 1. [Google Scholar] [CrossRef] [Green Version]
- Henningson, E.W.; Ahlberg, M.S. Evaluation of Microbiological Aerosol Samplers: A Review. J. Aerosol Sci. 1994, 25, 1459–1492. [Google Scholar] [CrossRef]
- Pepper, I.L.; Gerba, C.P. Aeromicrobiology; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Mathanlal, T.; Israel Nazarious, M.; Mantas-Nakhai, R.; Zorzano, M.-P.; Martin-Torres, J. ATMO-Vent: An Adapted Breathing Atmosphere for COVID-19 Patients. HardwareX 2020, 8, e00145. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Ravichandar, J.D.; Jain, S.; Griffin, D.W.; Yu, H.; Tan, Q.; Thissen, J.; Lusby, T.; Nicoll, P.; Shedler, S.; et al. Airborne Bacteria in Earth’s Lower Stratosphere Resemble Taxa Detected in the Troposphere: Results From a New NASA Aircraft Bioaerosol Collector (ABC). Front. Microbiol. 2018, 9, 1752. [Google Scholar] [CrossRef] [PubMed]
- Dommergue, A.; Amato, P.; Tignat-Perrier, R.; Magand, O.; Thollot, A.; Joly, M.; Bouvier, L.; Sellegri, K.; Vogel, T.; Sonke, J.E.; et al. Methods to Investigate the Global Atmospheric Microbiome. Front. Microbiol. 2019, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Hyde, P.; Mahalov, A. Contribution of Bioaerosols to Airborne Particulate Matter. J. Air Waste Manag. Assoc. 2020, 70, 71–77. [Google Scholar] [CrossRef]
- Xie, W.; Li, Y.; Bai, W.; Hou, J.; Ma, T.; Zeng, X.; Zhang, L.; An, T. The Source and Transport of Bioaerosols in the Air: A Review. Front. Environ. Sci. Eng. 2021, 15, 44. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.W. The Effect of Environmental Parameters on the Survival of Airborne Infectious Agents. J. R. Soc. Interface 2009, 6 (Suppl. S6), S737–S746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masago, K.; Fujita, S.; Oya, Y.; Takahashi, Y.; Matsushita, H.; Sasaki, E.; Kuroda, H. Comparison between Fluorimetry (Qubit) and Spectrophotometry (NanoDrop) in the Quantification of DNA and RNA Extracted from Frozen and FFPE Tissues from Lung Cancer Patients: A Real-World Use of Genomic Tests. Med. B Aires 2021, 57, 1375. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, B.; Lal, H.; Srivastava, A. Review of Bioaerosols in Indoor Environment with Special Reference to Sampling, Analysis and Control Mechanisms. Environ. Int. 2015, 85, 254–272. [Google Scholar] [CrossRef]
- Kravchik, T.; Gherman, U.; Laichter, Y. Aerosol Filtration by Fibrous and Membrane Filters. In Proceedings of the 19th Conference of the Israel Nuclear Societies, Colorado Springs, CO, USA, 16–18 May 1996; Volume 28, p. 6. [Google Scholar]
- Krekeler, C.; Ziehr, H.; Klein, J. Influence of Physicochemical Bacterial Surface Properties on Adsorption to Inorganic Porous Supports. Appl. Microbiol. Biotechnol. 1991, 35, 484–490. [Google Scholar] [CrossRef]
- Mackenzie, G.; Boa, A.N.; Diego-Taboada, A.; Atkin, S.L.; Sathyapalan, T. Sporopollenin, The Least Known Yet Toughest Natural Biopolymer. Front. Mater. 2015, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Müller, G.; Schubert, K.; Fiedler, F.; Bandlow, W. The CAMP-Binding Ectoprotein from Saccharomyces Cerevisiae Is Membrane-Anchored by Glycosyl-Phosphatidylinositol. J. Biol. Chem. 1992, 267, 25337–25346. [Google Scholar] [CrossRef] [PubMed]
- Kochkodan, V.; Tsarenko, S.; Potapchenko, N.; Kosinova, V.; Goncharuk, V. Adhesion of Microorganisms to Polymer Membranes: A Photobactericidal Effect of Surface Treatment with TiO2. Desalination 2008, 220, 380–385. [Google Scholar] [CrossRef]
- Giaouris, E.; Chapot-Chartier, M.-P.; Briandet, R. Surface Physicochemical Analysis of Natural Lactococcus Lactis Strains Reveals the Existence of Hydrophobic and Low Charged Strains with Altered Adhesive Properties. Int. J. Food Microbiol. 2009, 131, 2–9. [Google Scholar] [CrossRef]
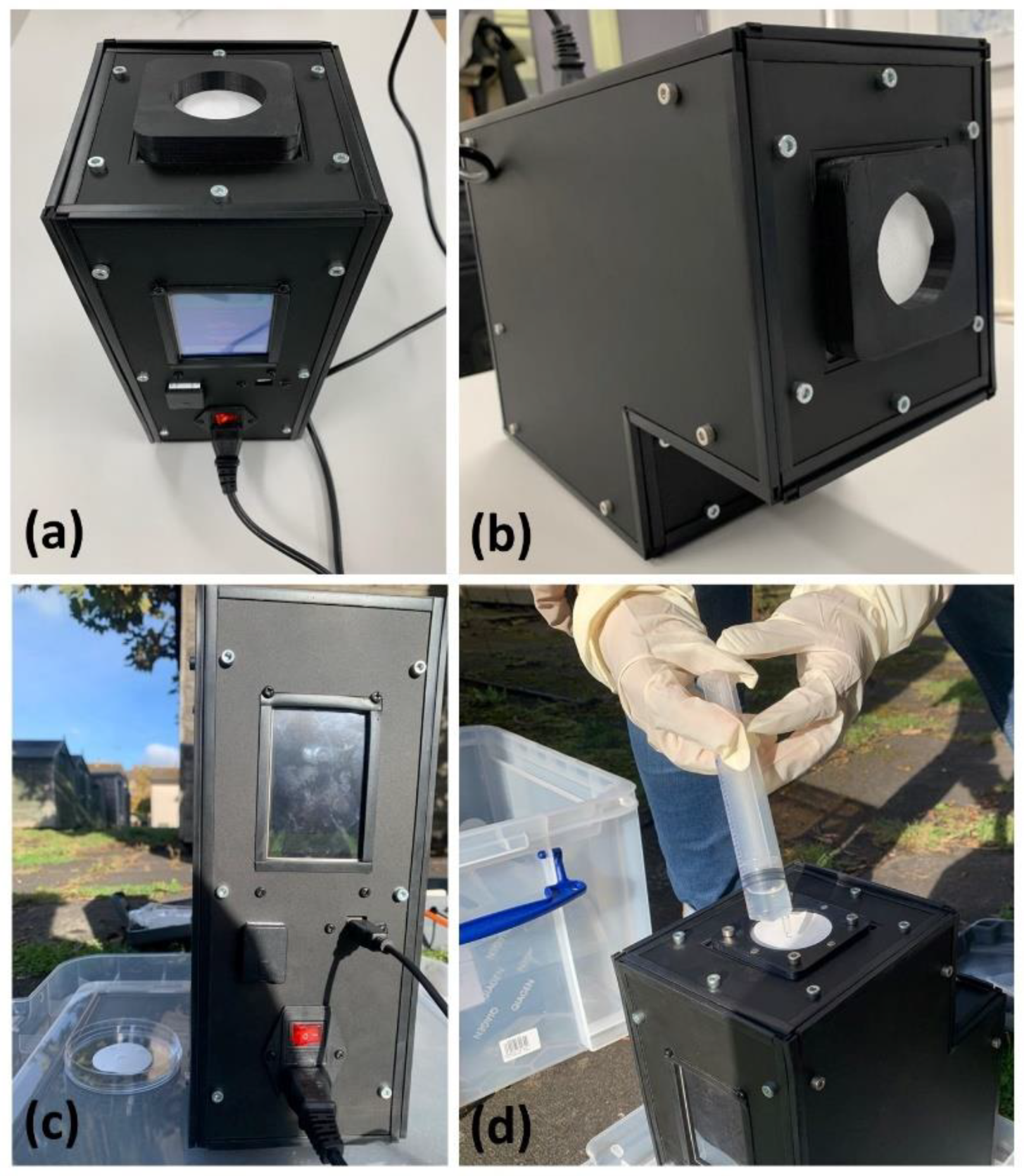
| Parameter | Range |
|---|---|
| Voltage | 220 V AC 50 Hz |
| Power | 50 W |
| Filter size | 47 mm diameter |
| Sampling time | 30 min–72 h |
| Sampling flow rate | 50–250 L/min (intervals of 50 L/min) |
| Temperature sensor characteristics | −40 °C to +85 °C; accuracy: ±1 °C; resolution: 0.010 °C |
| Humidity sensor characteristics | 5% to 95% RH; accuracy: ±3%; resolution: 0.7% |
| Pressure sensor characteristics | 300–1100 mbar; accuracy: ±2 mbar; resolution: 0.06 mbar |
| Filter | Product ID | Hydrophobic/Hydrophilic | Thickness | Air Flow Rate |
|---|---|---|---|---|
| PES | GPWP04700 | Hydrophilic | 160–185 µm | 6 L/min × cm2 |
| PTFE | FGLP04700 | Hydrophobic | 150 µm | 5 L/min × cm2 |
| PC | GTTP04700 | Hydrophilic | 7–22 µm | 0.007 L/min × cm2 |
| TissuQuartz | 513-0028 | Hydrophilic | 432 µm | 73 L/min × cm2 |
| Filter Type | Aerosol Source | Average T (°C) | Highest RH% | Average P (Bar) |
|---|---|---|---|---|
| Yeast supernatant | 20.91 | 61.78 | 1.002 | |
| PTFE | Soil supernatant | 20.86 | 65.92 | 1.005 |
| Negative control | 22.40 | 38.72 | 1.002 | |
| Yeast supernatant | 21.34 | 75.57 | 1.003 | |
| PES | Soil supernatant | 20.82 | 70.72 | 1.005 |
| Negative control | 22.57 | 40.02 | 1.002 | |
| Yeast supernatant | 21.68 | 83.68 | 1.003 | |
| PC | Soil supernatant | 20.68 | 79.39 | 1.009 |
| Negative control | 22.30 | 45.53 | 1.002 | |
| Yeast supernatant | 21.52 | 53.02 | 1.002 | |
| Quartz | Soil supernatant | 21.12 | 61.39 | 1.005 |
| Negative control | 22.24 | 39.42 | 1.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basapathi Raghavendra, J.; Mathanlal, T.; Zorzano, M.-P.; Martin-Torres, J. An Optimized Active Sampling Procedure for Aerobiological DNA Studies. Sensors 2023, 23, 2836. https://doi.org/10.3390/s23052836
Basapathi Raghavendra J, Mathanlal T, Zorzano M-P, Martin-Torres J. An Optimized Active Sampling Procedure for Aerobiological DNA Studies. Sensors. 2023; 23(5):2836. https://doi.org/10.3390/s23052836
Chicago/Turabian StyleBasapathi Raghavendra, Jyothi, Thasshwin Mathanlal, Maria-Paz Zorzano, and Javier Martin-Torres. 2023. "An Optimized Active Sampling Procedure for Aerobiological DNA Studies" Sensors 23, no. 5: 2836. https://doi.org/10.3390/s23052836






