Respiratory Rate Extraction from Neonatal Near-Infrared Spectroscopy Signals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
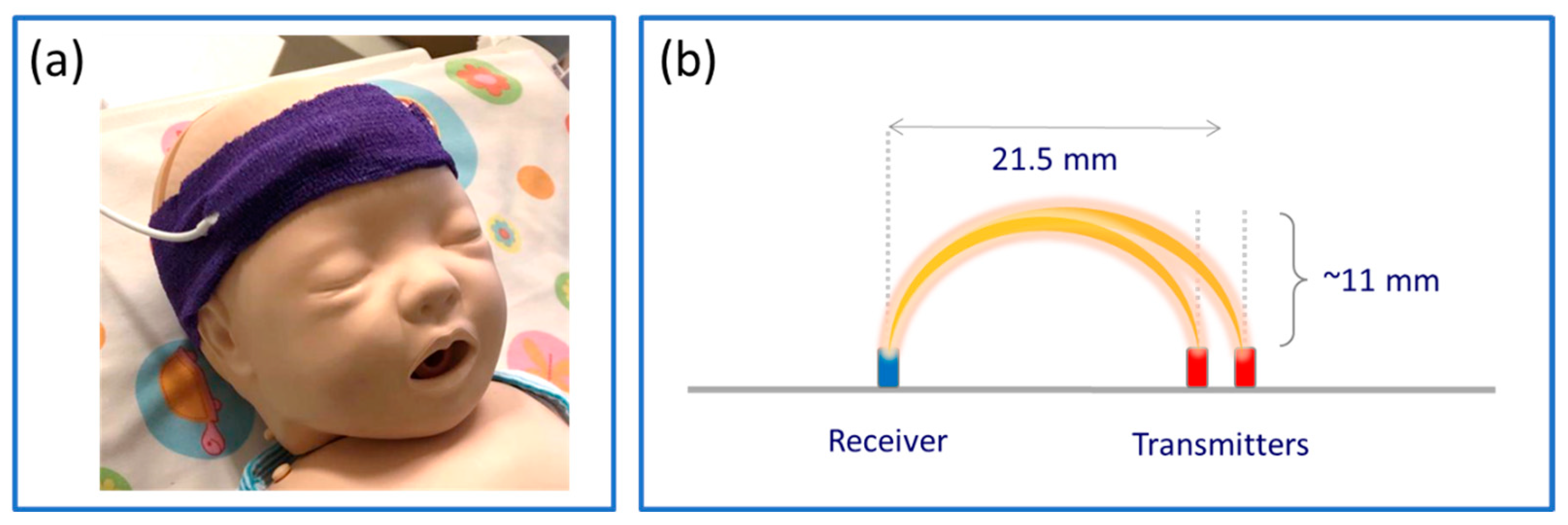
2.2. Data Acquisition
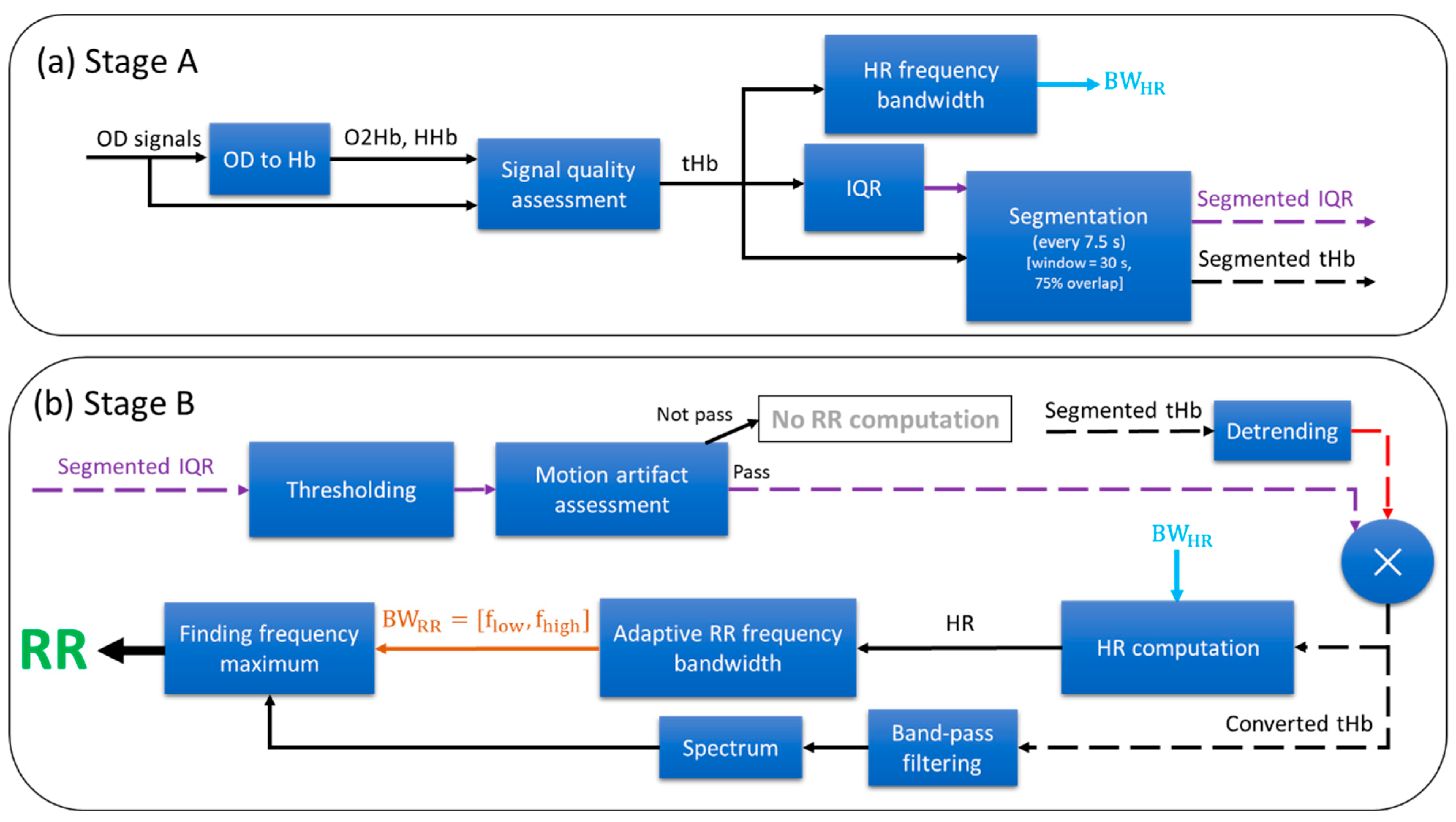
2.3. Respiratory Rate Extraction Algorithm
2.3.1. Preprocessing
2.3.2. HR Frequency Bandwidth
2.3.3. Interquartile Range (IQR)
2.3.4. Segmentation
2.3.5. Motion Artifact Assessment
2.3.6. HR Computation
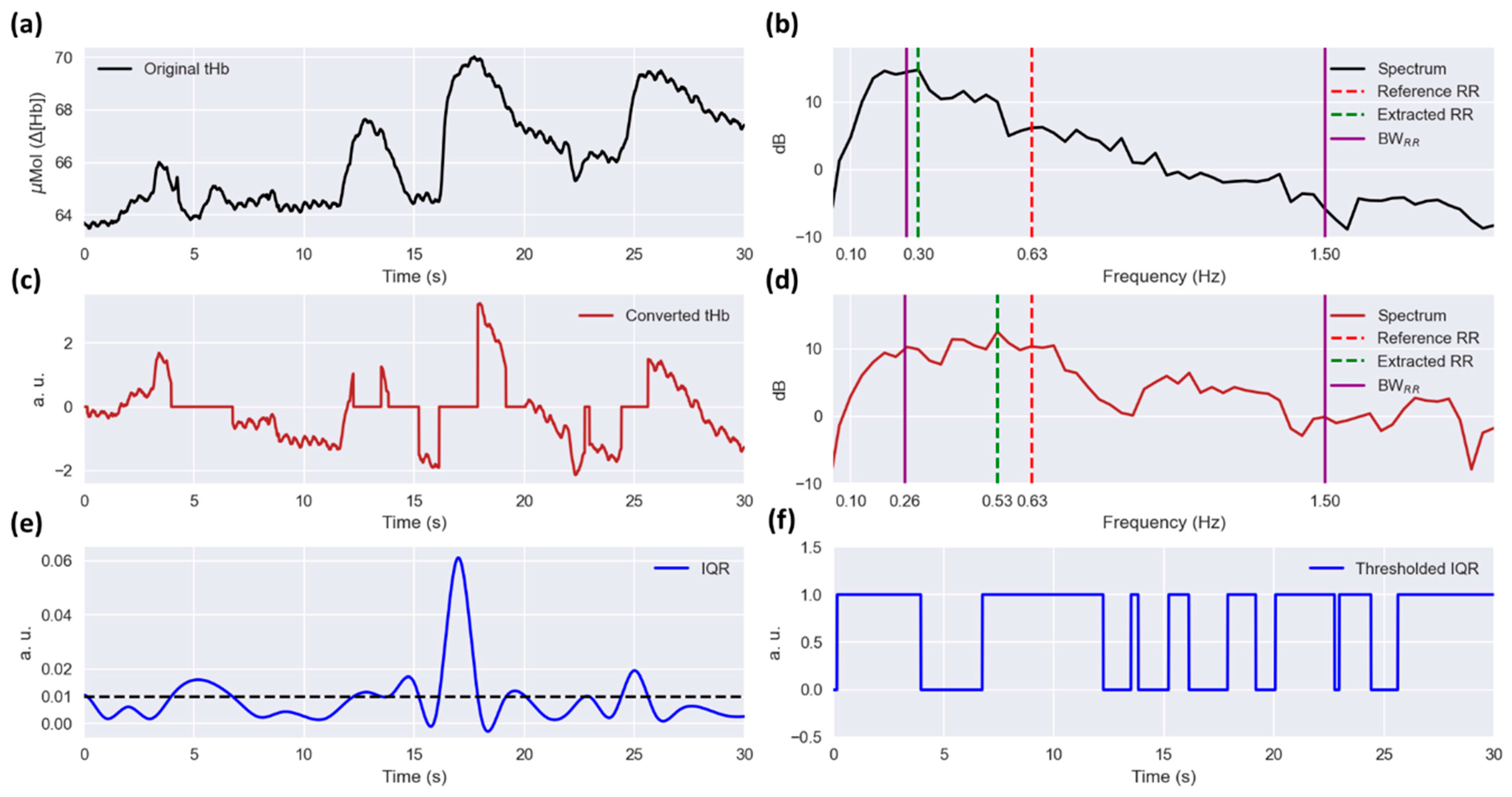
2.3.7. RR Computation
2.4. Algorithm Performance Assessment
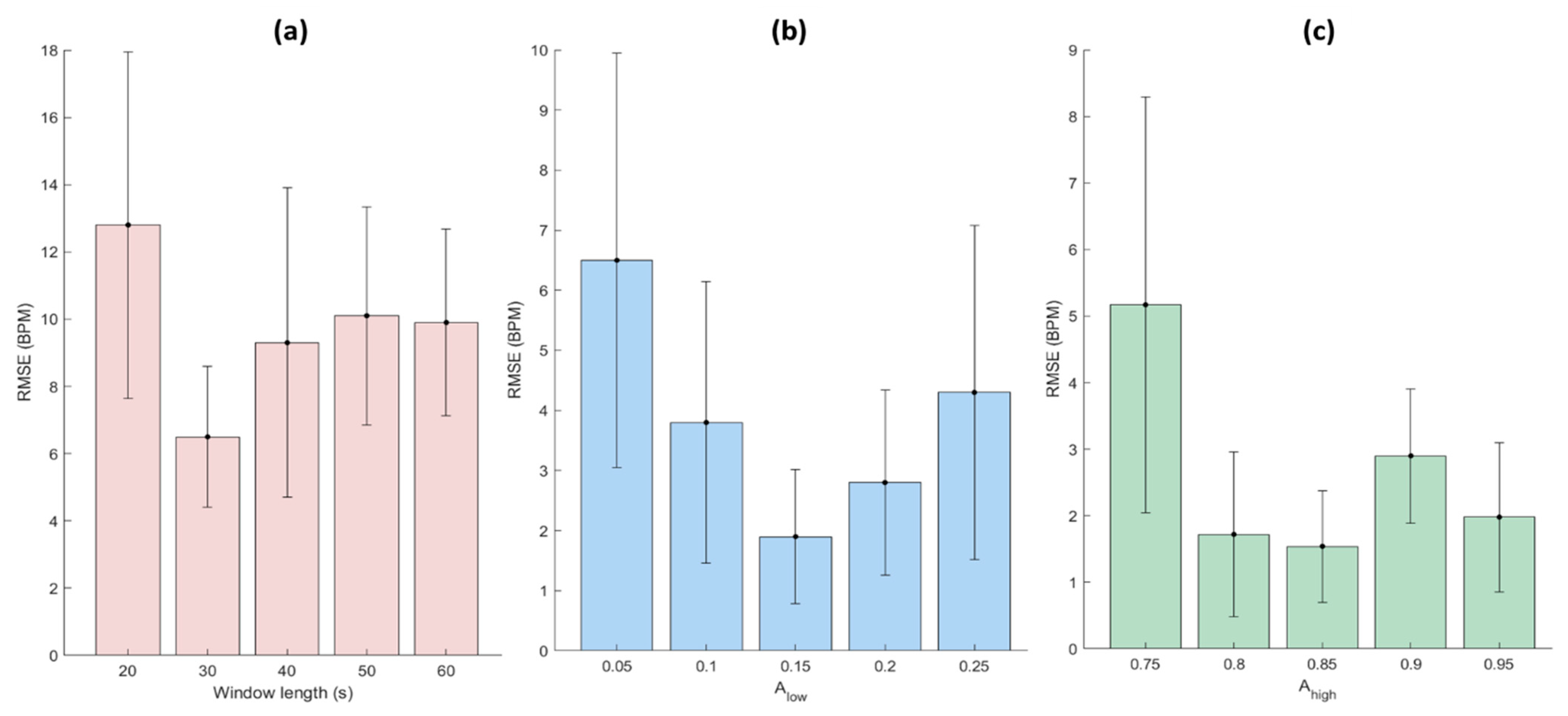
2.5. Optimization of the Parameters
3. Results
3.1. Optimization of the Proposed Algorithm’s Parameters
3.2. Results of the Proposed Algorithm
3.3. Comparison with the Existing Algorithms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fuerch, J.H.; Thio, M.; Halamek, L.P.; Liley, H.G.; Wyckoff, M.H.; Rabi, Y. Respiratory Function Monitoring during Neonatal Resuscitation: A Systematic Review. Resusc. Plus 2022, 12, 100327. [Google Scholar] [CrossRef]
- Fitzgerald, D.; van Asperen, P.; O’leary, P.; Feddema, P.; Leslie, G.; Arnold, J.; Sullivan, C. Sleep, Respiratory Rate, and Growth Hormone in Chronic Neonatal Lung Disease. Pediatr. Pulmonol. 1998, 26, 241–249. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Fairchild, K.D. Vital Signs as Physiomarkers of Neonatal Sepsis. Pediatr. Res. 2021, 91, 273–282. [Google Scholar] [CrossRef]
- Vavrinsky, E.; Esfahani, N.E.; Hausner, M.; Kuzma, A.; Rezo, V.; Donoval, M.; Kosnacova, H. The Current State of Optical Sensors in Medical Wearables. Biosensors 2022, 12, 217. [Google Scholar] [CrossRef]
- Lorato, I.; Stuijk, S.; Meftah, M.; Kommers, D.; Andriessen, P.; van Pul, C.; de Haan, G. Towards Continuous Camera-Based Respiration Monitoring in Infants. Sensors 2021, 21, 2268. [Google Scholar] [CrossRef]
- Reuter, S.; Moser, C.; Baack, M. Respiratory Distress in the Newborn. Pediatr. Rev. 2014, 35, 417–428. [Google Scholar] [CrossRef]
- Jenni, O.G.; Wolf, M.; Hengartner, M.; von Siebenthal, K.; Keel, M.; Bucher, H.U. Impact of Central, Obstructive and Mixed Apnea on Cerebral Hemodynamics in Preterm Infants. Neonatology 1996, 70, 91–100. [Google Scholar] [CrossRef]
- Horne, R.S.C.; Fung, A.C.H.; NcNeil, S.; Fyfe, K.L.; Odoi, A.; Wong, F.Y. The Longitudinal Effects of Persistent Apnea on Cerebral Oxygenation in Infants Born Preterm. J. Pediatr. 2017, 182, 79–84. [Google Scholar] [CrossRef]
- Payer, C.; Urlesberger, B.; Pauger, M.; Müller, W. Apnea Associated with Hypoxia in Preterm Infants: Impact on Cerebral Blood Volume. Brain Dev. 2003, 25, 25–31. [Google Scholar] [CrossRef]
- Yamamoto, A.; Yokoyama, N.; Yonetani, M.; Uetani, Y.; Nakamura, H.; Nakao, H. Evaluation of Change of Cerebral Circulation by SpO2 in Preterm Infants with Apneic Episodes Using near Infrared Spectroscopy. Pediatr. Int. 2003, 45, 661–664. [Google Scholar] [CrossRef]
- Coleman, J.; Ginsburg, A.S.; Macharia, W.M.; Ochieng, R.; Chomba, D.; Zhou, G.; Dunsmuir, D.; Karlen, W.; Ansermino, J.M. Assessment of Neonatal Respiratory Rate Variability. J. Clin. Monit. Comput. 2022, 36, 1869–1879. [Google Scholar] [CrossRef]
- Choi, S.H.; Lee, J.; Nam, S.K.; Jun, Y.H. Cerebral Oxygenation during Apnea in Preterm Infants: Effects of Accompanying Peripheral Oxygen Desaturation. Neonatal Med. 2021, 28, 14–21. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, T.; Li, G.; Li, T.; Inoue, Y. Wearable Sensor Systems for Infants. Sensors 2015, 15, 3721–3749. [Google Scholar] [CrossRef]
- Villringer, A.; Planck, J.; Hock, C.; Schleinkofer, L.; Dirnagl, U. Near Infrared Spectroscopy (NIRS): A New Tool to Study Hemodynamic Changes during Activation of Brain Function in Human Adults. Neurosci. Lett. 1993, 154, 101–104. [Google Scholar] [CrossRef]
- Sood, B.G.; McLaughlin, K.; Cortez, J. Near-Infrared Spectroscopy: Applications in Neonates. Semin. Fetal Neonatal Med. 2015, 20, 164–172. [Google Scholar] [CrossRef]
- Hansen, M.L.; Hyttel-Sørensen, S.; Jakobsen, J.C.; Gluud, C.; Kooi, E.M.W.; Mintzer, J.; de Boode, W.P.; Fumagalli, M.; Alarcon, A.; Alderliesten, T.; et al. Cerebral Near-Infrared Spectroscopy Monitoring (NIRS) in Children and Adults: A Systematic Review with Meta-Analysis. Pediatr. Res. 2022, 1–12. [Google Scholar] [CrossRef]
- Zhang, F.; Cheong, D.; Khan, A.F.; Chen, Y.; Ding, L.; Yuan, H. Correcting Physiological Noise in Whole-Head Functional near-Infrared Spectroscopy. J. Neurosci. Methods 2021, 360, 109262. [Google Scholar] [CrossRef]
- Costa, F.G.; Hakimi, N.; Van Bel, F. Neuroprotection of the Perinatal Brain by Early Information of Cerebral Oxygenation and Perfusion Patterns. Int. J. Mol. Sci. 2021, 22, 5389. [Google Scholar] [CrossRef]
- Hakimi Seyed Kamaledin Setarehdan Naser Hakimi, N.; Kamaledin Setarehdan, S.; Hakimi, N. Stress Assessment by Means of Heart Rate Derived from Functional Near-Infrared Spectroscopy. J. Biomed. Opt. 2018, 23, 115001. [Google Scholar] [CrossRef]
- Hakimi, N.; Jodeiri, A.; Mirbagheri, M.; Setarehdan, S.K. Proposing a Convolutional Neural Network for Stress Assessment by Means of Derived Heart Rate from Functional near Infrared Spectroscopy. Comput. Biol. Med. 2020, 121, 103810. [Google Scholar] [CrossRef]
- Hakimi, N.; Shahbakhti, M.; Sappia, S.; Horschig, J.M.; Bronkhorst, M.; Floor-Westerdijk, M.; Valenza, G.; Dudink, J.; Colier, W.N.J.M. Estimation of Respiratory Rate from Functional Near-Infrared Spectroscopy (FNIRS): A New Perspective on Respiratory Interference. Biosensors 2022, 12, 1170. [Google Scholar] [CrossRef]
- Reddy, P.; Izzetoglu, M.; Shewokis, P.A.; Sangobowale, M.; Diaz-Arrastia, R.; Izzetoglu, K. Evaluation of FNIRS Signal Components Elicited by Cognitive and Hypercapnic Stimuli. Sci. Rep. 2021, 11, 23457. [Google Scholar] [CrossRef]
- Iqbal, T.; Elahi, A.; Ganly, S.; Wijns, W.; Shahzad, A. Photoplethysmography-Based Respiratory Rate Estimation Algorithm for Health Monitoring Applications. J. Med. Biol. Eng. 2022, 42, 242–252. [Google Scholar] [CrossRef]
- Tong, Y.; Lindsey, K.P.; Frederick, B.D. Partitioning of Physiological Noise Signals in the Brain with Concurrent Near-Infrared Spectroscopy and FMRI. J. Cereb. Blood Flow Metab. 2011, 31, 2352–2362. [Google Scholar] [CrossRef]
- Kuller, J.M.M. Skin Breakdown: Risk Factors, Prevention, and Treatment. Newborn Infant Nurs. Rev. 2001, 1, 35–42. [Google Scholar] [CrossRef]
- Murray, J.S.; Noonan, C.; Quigley, S.; Curley, M.A.Q. Medical Device-Related Hospital-Acquired Pressure Ulcers in Children: An Integrative Review. J. Pediatr. Nurs. 2013, 28, 585–595. [Google Scholar] [CrossRef]
- Anand, K.J.S.; Scalzo, F.M. Can Adverse Neonatal Experiences Alter Brain Development and Subsequent Behavior? Biol. Neonate 2000, 77, 69–82. [Google Scholar] [CrossRef]
- Afsar, F.S. Skin Care for Preterm and Term Neonates. Clin. Exp. Dermatol. 2009, 34, 855–858. [Google Scholar] [CrossRef]
- Ludington-Hoe, S.M.; Anderson, G.C.; Swinth, J.Y.; Thompson, C.; Hadeed, A.J. Randomized Controlled Trial of Kangaroo Care: Cardiorespiratory and Thermal Effects on Healthy Preterm Infants. Neonatal Netw. 2004, 23, 39–48. [Google Scholar] [CrossRef]
- Pereira, C.B.; Czaplik, M.; Blazek, V.; Leonhardt, S.; Teichmann, D. Monitoring of Cardiorespiratory Signals Using Thermal Imaging: A Pilot Study on Healthy Human Subjects. Sensors 2018, 18, 1541. [Google Scholar] [CrossRef]
- Quaresima, V.; Bisconti, S.; Ferrari, M. A Brief Review on the Use of Functional Near-Infrared Spectroscopy (FNIRS) for Language Imaging Studies in Human Newborns and Adults. Brain Lang. 2012, 121, 79–89. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Delpy, D.T.; Cope, M.; Van Der Zee, P.; Arridge, S.; Wray, S.; Wyatt, J. Estimation of Optical Pathlength through Tissue from Direct Time of Flight Measurement. Phys. Med. Biol. 1988, 33, 1433. [Google Scholar] [CrossRef]
- Sappia, M.S.; Sappia, M.S.; Sappia, M.S.; Hakimi, N.; Hakimi, N.; Hakimi, N.; Colier, W.N.J.M.; Horschig, J.M. Signal Quality Index: An Algorithm for Quantitative Assessment of Functional near Infrared Spectroscopy Signal Quality. Biomed. Opt. Express 2020, 11, 6732–6754. [Google Scholar] [CrossRef]
- Treitel, S. Spectral Analysis for Physical Applications: Multitaper and Conventional Univariate Techniques. Am. Sci. 1995, 83, 195–197. [Google Scholar]
- Slepian, D. Prolate Spheroidal Wave Functions, Fourier Analysis, and Uncertainty—V: The Discrete Case. Bell Syst. Tech. J. 1978, 57, 1371–1430. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Measurement in Medicine: The Analysis of Method Comparison Studies. J. R. Stat. Soc. Ser. D (Stat.) 1983, 32, 307–317. [Google Scholar] [CrossRef]
- Pollonini, L.; Olds, C.; Abaya, H.; Bortfeld, H.; Beauchamp, M.S.; Oghalai, J.S. Auditory Cortex Activation to Natural Speech and Simulated Cochlear Implant Speech Measured with Functional Near-Infrared Spectroscopy. Hear. Res. 2014, 309, 84–93. [Google Scholar] [CrossRef]
- Sappia, M.S.; Hakimi, N.; Svinkunaite, L.; Alderliesten, T.; Horschig, J.M.; Colier, W.N.J.M. FNIRS Signal Quality Estimation by Means of a Machine Learning Algorithm Trained on Morphological and Temporal Features; SPIE: Bellingham, WA, USA, 2021; Volume 11638, pp. 29–39. [Google Scholar] [CrossRef]
- Holper, L.; Seifritz, E.; Scholkmann, F. Short-Term Pulse Rate Variability Is Better Characterized by Functional near-Infrared Spectroscopy than by Photoplethysmography. J. Biomed. Opt. 2016, 21, 091308. [Google Scholar] [CrossRef]
- Mirbagheri, M.; Hakimi, N.; Ebrahimzadeh, E.; Setarehdan, S.K. Quality Analysis of Heart Rate Derived from Functional Near-Infrared Spectroscopy in Stress Assessment. Inform. Med. Unlocked 2020, 18, 100286. [Google Scholar] [CrossRef]
- Perdue, K.L.; Westerlund, A.; Mccormick, S.A.; Nelson, C.A.; Iii, C.A.N. Extraction of Heart Rate from Functional Near-Infrared Spectroscopy in Infants. J. Biomed. Opt. 2014, 19, 067010. [Google Scholar] [CrossRef]
- Daw, W.; Kaur, R.; Delaney, M.; Elphick, H. Respiratory Rate Is an Early Predictor of Clinical Deterioration in Children. Pediatr. Pulmonol. 2020, 55, 2041–2049. [Google Scholar] [CrossRef]
- Braun, S.R. Respiratory Rate and Pattern. In Clinical Methods: The History, Physical, and Laboratory Examinations; Butterworth Publishers: Oxford, UK, 1990. [Google Scholar]
- Cretikos, M.A.; Bellomo, R.; Hillman, K.; Chen, J.; Finfer, S.; Flabouris, A. Respiratory Rate: The Neglected Vital Sign. Med. J. Aust. 2008, 188, 657–659. [Google Scholar] [CrossRef]
- Elliott, M.; Coventry, A. Critical Care: The Eight Vital Signs of Patient Monitoring. Br. J. Nurs. 2013, 21, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.H. The Epidemiology of Respiratory Failure in Neonates Born at an Estimated Gestational Age of 34 Weeks or More. J. Perinatol. 2004, 25, 251–257. [Google Scholar] [CrossRef]
- Szabo, S.M.; Gooch, K.L.; Korol, E.E.; Bradt, P.; Vo, P.; Levy, A.R. Respiratory Distress Syndrome at Birth Is a Risk Factor for Hospitalization for Lower Respiratory Tract Infections in Infancy. Pediatr. Infect. Dis. J. 2012, 31, 1245–1251. [Google Scholar] [CrossRef]
- Liu, J.; Sorantin, E. Neonatal Respiratory Distress Syndrome. In Neonatal Lung Ultrasonography; Springer: Dordrecht, The Netherlands, 2020; pp. 17–39. [Google Scholar] [CrossRef]
- Ahmed, W.; Veluthandath, A.V.; Rowe, D.J.; Madsen, J.; Clark, H.W.; Postle, A.D.; Wilkinson, J.S.; Murugan, G.S. Prediction of Neonatal Respiratory Distress Biomarker Concentration by Application of Machine Learning to Mid-Infrared Spectra. Sensors 2022, 22, 1744. [Google Scholar] [CrossRef]
- Dasgupta, S.J.; Gill, A.B. Hypotension in the Very Low Birthweight Infant: The Old, the New, and the Uncertain. Arch. Dis. Child. Fetal Neonatal Ed. 2003, 88, F450–F454. [Google Scholar] [CrossRef]
- Petrova, A.; Mehta, R. Near-Infrared Spectroscopy in the Detection of Regional Tissue Oxygenation during Hypoxic Events in Preterm Infants Undergoing Critical Care. Pediatr. Crit. Care Med. 2006, 7, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Owen-Reece, H.; Elwell, C.E.; Goldstone, J.; Smith, M.; Delpy, D.T.; Wyatt, J.S. Investigation of the Effects of Hypocapnia upon Cerebral Haemodynamics in Normal Volunteers and Anaesthetised Subjects by near Infrared Spectroscopy (NIRS). Adv. Exp. Med. Biol. 1994, 361, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, L.; Habre, W.; Pichon, I.; Fodor, G.H. Effect of Permissive Mild Hypercapnia on Cerebral Vasoreactivity in Infants: A Randomized Controlled Crossover Trial. Anesth. Analg. 2021, 133, 976–983. [Google Scholar] [CrossRef]
- Sandal, G.; Oguz, S.S.; Erdeve, O.; Akar, M.; Uras, N.; Dilmen, U. Assessment of Red Blood Cell Transfusion and Transfusion Duration on Cerebral and Mesenteric Oxygenation Using Near-Infrared Spectroscopy in Preterm Infants with Symptomatic Anemia. Transfusion 2014, 54, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Korček, P.; Straňák, Z.; Širc, J.; Naulaers, G. The Role of Near-Infrared Spectroscopy Monitoring in Preterm Infants. J. Perinatol. 2017, 37, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Senese, V.P. Functional Near-Infrared Spectroscopy Is a Useful Tool for Multi-Perspective Psychobiological Study of Neurophysiological Correlates of Parenting Behaviour. Eur. J. Neurosci. 2023, 57, 258–284. [Google Scholar] [CrossRef]
- Nguyen, T.; Abney, D.H.; Salamander, D.; Bertenthal, B.I.; Hoehl, S. Proximity and Touch Are Associated with Neural but Not Physiological Synchrony in Naturalistic Mother-Infant Interactions. Neuroimage 2021, 244, 118599. [Google Scholar] [CrossRef]

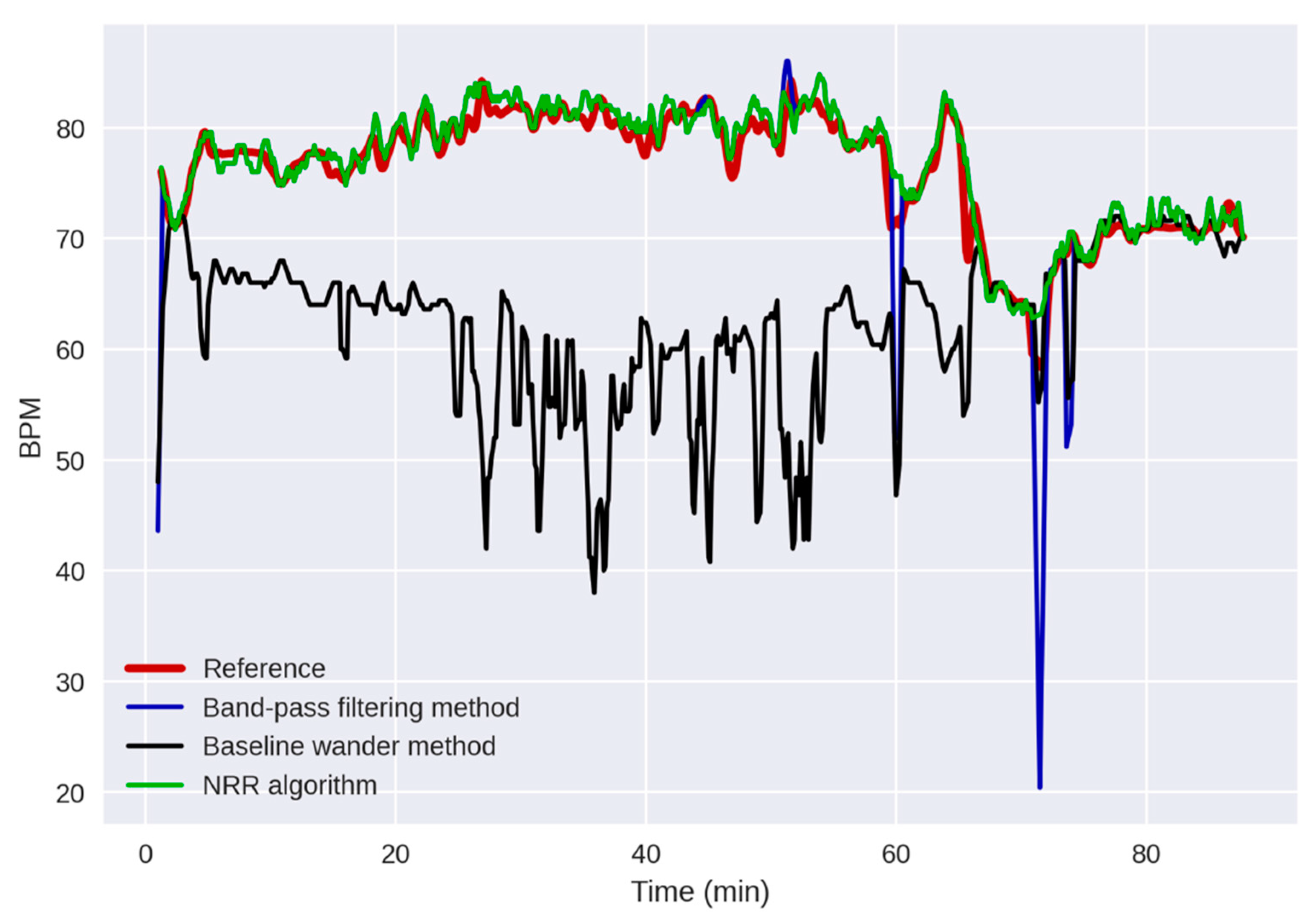

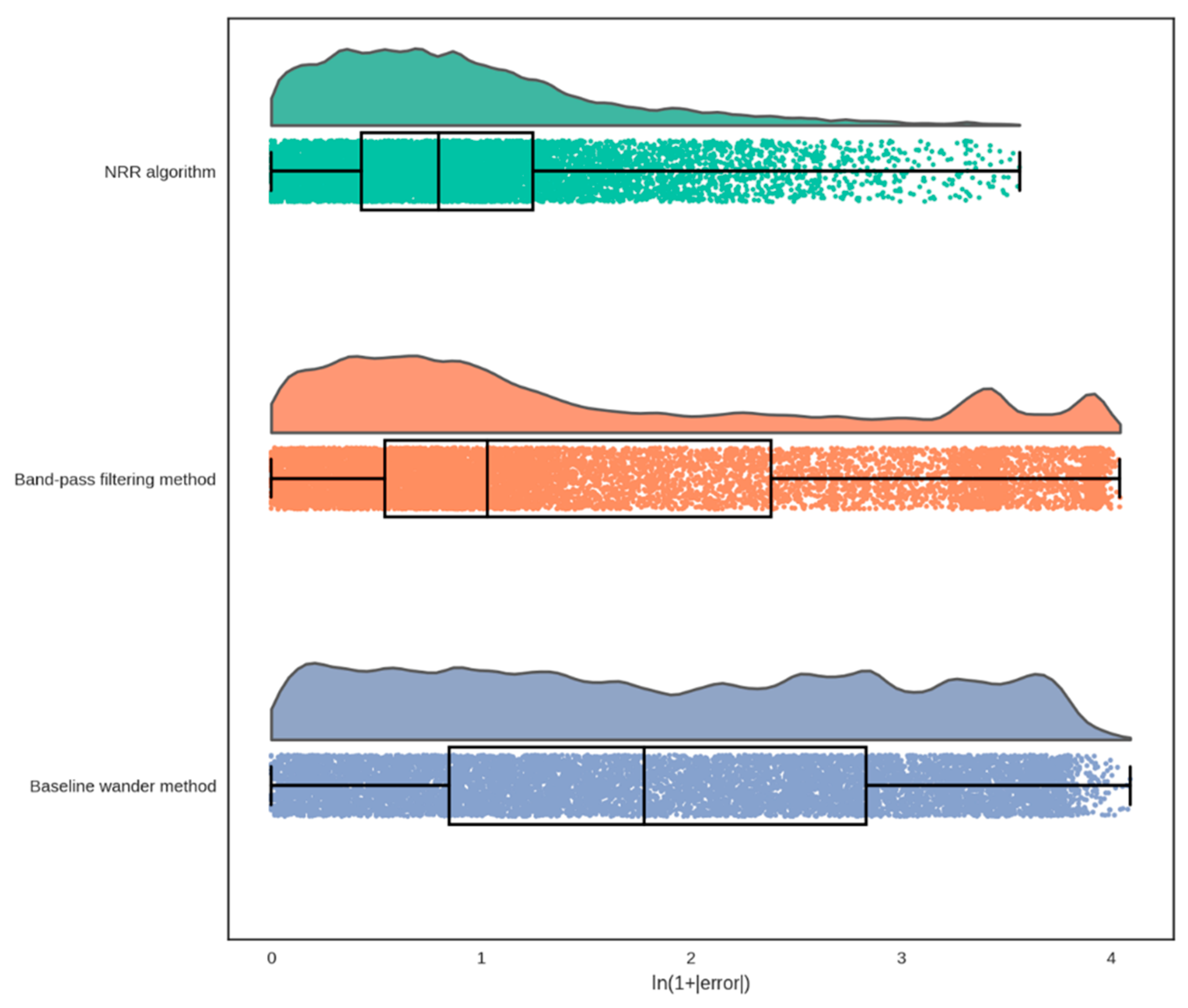
| Measurement | ME 1 (BPM 2) | RMSE 3 (BPM) | LoA 4 (BPM) | Pearson’s r (%) | Included Segments (%) |
|---|---|---|---|---|---|
| 1 | 2.4 | 5.9 | 10.6 | 76.1 | 92.1 |
| 2 | 0.2 | 1.3 | 2.5 | 83.8 | 99.8 |
| 3 | 0.3 | 1.7 | 3.2 | 94.4 | 99.9 |
| 4 | 0.4 | 8.1 | 15.9 | 72.9 | 79.2 |
| 5 | −0.6 | 1.5 | 2.6 | 96.7 | 99.0 |
| 6 | 0.9 | 2.0 | 3.4 | 84.6 | 100 |
| 7 | 2.7 | 5.1 | 8.4 | 48.4 | 99.1 |
| 8 | 0.3 | 3.8 | 7.5 | 85.5 | 95.5 |
| 9 | −0.1 | 2.0 | 4.0 | 90.4 | 96.2 |
| 10 | −0.5 | 1.5 | 2.7 | 95.7 | 94.5 |
| 11 | −0.1 | 1.8 | 3.6 | 87.5 | 98.8 |
| 12 | 0.0 | 2.5 | 4.9 | 93.1 | 87.0 |
| 13 | 1.1 | 3.5 | 6.5 | 89.6 | 96.0 |
| 14 | 0.8 | 4.4 | 8.6 | 93.6 | 96.5 |
| 15 | 8.1 | 12.2 | 18.1 | 71.4 | 69.6 |
| 16 | 1.0 | 2.9 | 5.7 | 87.6 | 97.4 |
| Average | 1.1 | 3.8 | 6.7 | 84.5 | 93.8 |
| Std 5 | 2.1 | 3.0 | 4.7 | 12.3 | 8.5 |
| Algorithm | ME 1 (BPM 2) | RMSE 3 (BPM) | LoA 4 (BPM) | Pearson’s r (%) |
|---|---|---|---|---|
| NRR | 1.1 ± 2.1 | 3.8 ± 3.0 | 6.7 ± 4.7 | 84.5 ± 12.3 |
| BPF | −1.4 ± 11.6 | 10.0 ± 10.7 | 14.0 ± 11.1 | 70.8 ± 15.0 |
| BW | 8.4 ± 14.1 | 12.6 ± 12.2 | 11.4 ± 5.8 | 51.1 ± 26.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hakimi, N.; Shahbakhti, M.; Horschig, J.M.; Alderliesten, T.; Van Bel, F.; Colier, W.N.J.M.; Dudink, J. Respiratory Rate Extraction from Neonatal Near-Infrared Spectroscopy Signals. Sensors 2023, 23, 4487. https://doi.org/10.3390/s23094487
Hakimi N, Shahbakhti M, Horschig JM, Alderliesten T, Van Bel F, Colier WNJM, Dudink J. Respiratory Rate Extraction from Neonatal Near-Infrared Spectroscopy Signals. Sensors. 2023; 23(9):4487. https://doi.org/10.3390/s23094487
Chicago/Turabian StyleHakimi, Naser, Mohammad Shahbakhti, Jörn M. Horschig, Thomas Alderliesten, Frank Van Bel, Willy N. J. M. Colier, and Jeroen Dudink. 2023. "Respiratory Rate Extraction from Neonatal Near-Infrared Spectroscopy Signals" Sensors 23, no. 9: 4487. https://doi.org/10.3390/s23094487





