Application of Data Fusion in Traditional Chinese Medicine: A Review
Abstract
:1. Introduction
2. Data Fusion Technology
2.1. Introduction to Data Fusion Technology
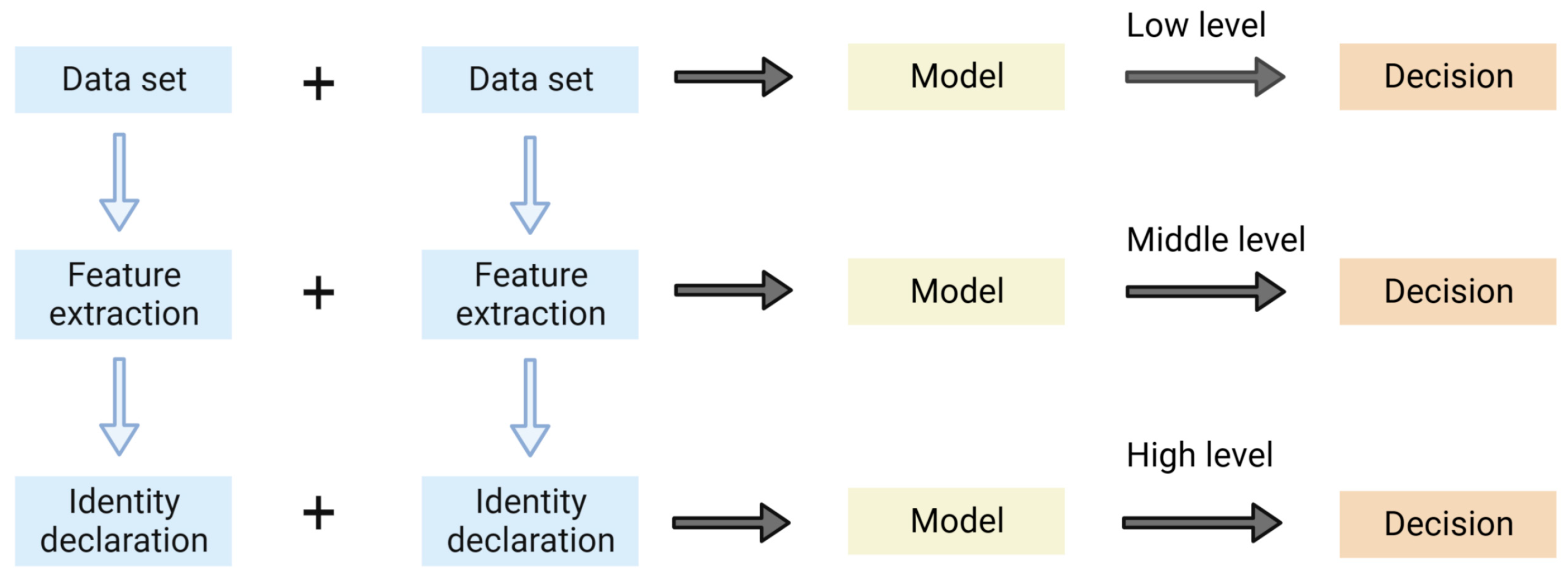
2.2. Data Fusion Strategy
2.3. Pre-Processing
3. Traditional Chinese Medicine and Data Analysis
3.1. Problems and Challenges Facing Traditional Chinese Medicine
3.2. Data Analysis Techniques and Traditional Chinese Medicine
3.3. Model Validation
4. Application of Data Fusion Technology in Traditional Chinese Medicine
4.1. Data Fusion for Different Spectroscopy Data
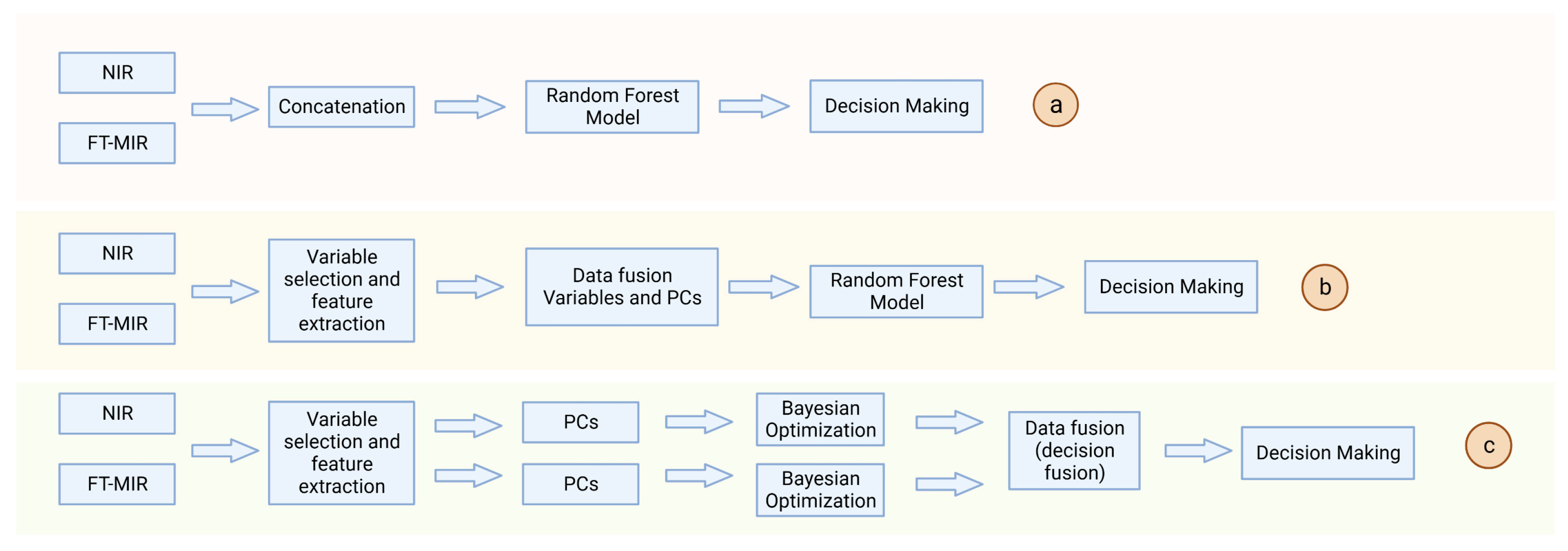
4.1.1. Data Fusion for Infrared Spectroscopy Data
4.1.2. Data Fusion for Other Spectroscopy Data
4.2. Data Fusion for Spectroscopy Data and Other Techniques Data
| TCM | Analytical Techniques | Chemometrics | Fusion Level | Ref. |
|---|---|---|---|---|
| Gentiana rigescens | IR, UV | PLS-DA, SVM | mid | [88] |
| Leccinum rugosiceps | IR, UV | PLS-DA, SVM | mid | [89] |
| Chinese herbal injection | NIRS, UVS | PLS, UVPLS | low, mid | [80] |
| cultivated Macrohyporia cocos | ATR-FTIR, UFLC | PLS-DA, PLSR | - | [90] |
| Wolfiporia cocos | FT-NIR, FT-MIR | PLS-DA, SVM PCA, HCA | low, mid | [91] |
| wildParis polyphylla Smith var. yunnanensis | FT-MIR, UV-Vis | SVM-GS, RF | low, mid, high | [92] |
| Polygonatum kingianum | ATR-FTIR, UV-Vis | PLS-DA, PCA, HCA | Low, mid, high | [93] |
| Dendrobium huoshanense | nano-effect NIR nano-effect MIR | PLS-DA, OPLS-DA | mid | [75] |
| Radix puerariae | NIR, UV | PLSR | low | [94] |
| Lonicera japonica and Artemisia annua | NIR, MIR | C-PLS, SO-PLS | - | [95] |
| Dendrobium Species | NIR, UV | SVM, PLS-DA | low, mid, high | [96] |
| Coptidis Rhizoma | NIR, MIR | PLS-DA, PLSR | low, mid, high | [97] |
| Radix Astragali | Vis-NIR, NIR | PCA, CNN, SVM, LR | - | [98] |
| Boletus bainiugan | FT-NIR, FT-MIR | PLS-DA, SVM 3DCOS-ResNet | low, mid | [99] |
| Honey | Gas sensors, Liquid sensors | LDA | low | [100] |
| Macrohyporia cocos | FTIR, LC | PLS-DA | low, high | [101] |
| Macrohyporia cocos | FTIR, HPLC | PLS-DA, PCA | mid | [102] |
| Swertia leducii | FTIR, UPLC | HCA, RF | low, mid | [83] |
| sulfur-Ophiopogonis Radix | NIR, UHPLC-HRMS | PCA, PLS-LDA | mid | [86] |
| Curcumae Rhizoma | FT-NIR, E-nose, colormeter | PLS-DA | mid | [103] |
4.3. Data Fusion for Mass Spectrometry and Chromatography
4.3.1. Data Fusion for Mass Spectrometry Data
4.3.2. Data Fusion for Chromatography Data
4.4. Data Fusion for Sensor Data
5. Comprehensive Evaluation of Data Fusion
6. Conclusions and Future Outlooks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.-Y.; Xie, Y.; Zhou, H.; Liu, L. Contribution of Traditional Chinese Medicine to the Treatment of COVID-19. Phytomedicine 2021, 85, 153279. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, Y.; He, Y.; Hou, W.; Liao, M.; Liu, C. Quality Markers of Traditional Chinese Medicine: Concept, Progress, and Perspective. Engineering 2019, 5, 888–894. [Google Scholar] [CrossRef]
- Tahir, H.E.; Arslan, M.; Komla Mahunu, G.; Adam Mariod, A.; Hashim, S.; Xiaobo, Z.; Jiyong, S.; El-Seedi, H.R.; Musa, T.H. The Use of Analytical Techniques Coupled with Chemometrics for Tracing the Geographical Origin of Oils: A Systematic Review (2013–2020). Food Chem. 2022, 366, 130633. [Google Scholar] [CrossRef] [PubMed]
- Doeswijk, T.G.; Smilde, A.K.; Hageman, J.A.; Westerhuis, J.A.; Van Eeuwijk, F.A. On the Increase of Predictive Performance with High-Level Data Fusion. Anal. Chim. Acta 2011, 705, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Yu, L.; Wang, C.; Zhong, S.; Gu, R. Quality assessment of traditional Chinese medicine based on data fusion combined with machine learning: A review. Crit. Rev. Anal. Chem. 2023, 26, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Brierley, N.; Tippetts, T.; Cawley, P. Data Fusion for Automated Non-Destructive Inspection. Proc. R. Soc. A 2014, 470, 20140167. [Google Scholar] [CrossRef]
- Lorenz, S.; Seidel, P.; Ghamisi, P.; Zimmermann, R.; Tusa, L.; Khodadadzadeh, M.; Contreras, I.C.; Gloaguen, R. Multi-Sensor Spectral Imaging of Geological Samples: A Data Fusion Approach Using Spatio-Spectral Feature Extraction. Sensors 2019, 19, 2787. [Google Scholar] [CrossRef]
- Chen, J.; Li, D.; Huang, R.; Chen, Z.; Li, W. Aero-Engine Remaining Useful Life Prediction Method with Self-Adaptive Multimodal Data Fusion and Cluster-Ensemble Transfer Regression. Reliab. Eng. Syst. Saf. 2023, 234, 109151. [Google Scholar] [CrossRef]
- Liu, H.; Gao, C.; Wang, X.; Dou, Z.; Zhang, W. Aeroelastic Prediction in Transonic Buffeting Flow with Data Fusion Method. Aerosp. Sci. Technol. 2023, 133, 108097. [Google Scholar] [CrossRef]
- Yan, H.; Zuo, H.; Sun, J.; Zhou, D.; Wang, H. Two-Stage Degradation Assessment and Prediction Method for Aircraft Engine Based on Data Fusion. Int. J. Aerosp. Eng. 2021, 2021, 1–16. [Google Scholar] [CrossRef]
- Chmielewski, M.; Kukiełka, M.; Pieczonka, P.; Gutowski, T. Methods and Analytical Tools for Assessing Tactical Situation in Military Operations Using Potential Approach and Sensor Data Fusion. Procedia Manuf. 2020, 44, 559–566. [Google Scholar] [CrossRef]
- Barrile, V.; Simonetti, S.; Citroni, R.; Fotia, A.; Bilotta, G. Experimenting Agriculture 4.0 with Sensors: A Data Fusion Approach between Remote Sensing, UAVs and Self-Driving Tractors. Sensors 2022, 22, 7910. [Google Scholar] [CrossRef] [PubMed]
- Steyaert, S.; Pizurica, M.; Nagaraj, D.; Khandelwal, P.; Hernandez-Boussard, T.; Gentles, A.J.; Gevaert, O. Multimodal Data Fusion for Cancer Biomarker Discovery with Deep Learning. Nat. Mach. Intell. 2023, 5, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Zhong, J.; Xue, R.; Su, W.; Kong, Y.; Fei, Y.; Ma, J.; Wang, Y.; Mi, L. Early Detection of Cervical Cancer by Fluorescence Lifetime Imaging Microscopy Combined with Unsupervised Machine Learning. IJMS 2022, 23, 11476. [Google Scholar] [CrossRef] [PubMed]
- Obisesan, K.A.; Jiménez-Carvelo, A.M.; Cuadros-Rodriguez, L.; Ruisánchez, I.; Callao, M.P. HPLC-UV and HPLC-CAD Chromatographic Data Fusion for the Authentication of the Geographical Origin of Palm Oil. Talanta 2017, 170, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Azcarate, S.M.; Ríos-Reina, R.; Amigo, J.M.; Goicoechea, H.C. Data Handling in Data Fusion: Methodologies and Applications. TrAC Trends Anal. Chem. 2021, 143, 116355. [Google Scholar] [CrossRef]
- Firmani, P.; Vitale, R.; Ruckebusch, C.; Marini, F. ANOVA-Simultaneous Component Analysis Modelling of Low-Level-Fused Spectroscopic Data: A Food Chemistry Case-Study. Anal. Chim. Acta 2020, 1125, 308–314. [Google Scholar] [CrossRef]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New Data Preprocessing Trends Based on Ensemble of Multiple Preprocessing Techniques. TrAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar] [CrossRef]
- Haddi, Z.; Mabrouk, S.; Bougrini, M.; Tahri, K.; Sghaier, K.; Barhoumi, H.; El Bari, N.; Maaref, A.; Jaffrezic-Renault, N.; Bouchikhi, B. E-Nose and e-Tongue Combination for Improved Recognition of Fruit Juice Samples. Food Chem. 2014, 150, 246–253. [Google Scholar] [CrossRef]
- Haddi, Z.; Boughrini, M.; Ihlou, S.; Amari, A.; Mabrouk, S.; Barhoumi, H.; Maaref, A.; El Bari, N.; Llobet, E.; Jaffrezic-Renault, N.; et al. Geographical Classification of Virgin Olive Oils by Combining the Electronic Nose and Tongue. In Proceedings of the 2012 IEEE Sensors Proceedings, Taipei, Taiwan, 28–31 October 2012; pp. 785–788. [Google Scholar]
- Haddi, Z.; Alami, H.; El Bari, N.; Tounsi, M.; Barhoumi, H.; Maaref, A.; Jaffrezic-Renault, N.; Bouchikhi, B. Electronic Nose and Tongue Combination for Improved Classification of Moroccan Virgin Olive Oil Profiles. Food Res. Int. 2013, 54, 1488–1498. [Google Scholar] [CrossRef]
- Biancolillo, A.; Bucci, R.; Magrì, A.L.; Magrì, A.D.; Marini, F. Data-Fusion for Multiplatform Characterization of an Italian Craft Beer Aimed at Its Authentication. Anal. Chim. Acta 2014, 820, 23–31. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y. Synergistic Strategy for the Geographical Traceability of Wild Boletus Tomentipes by Means of Data Fusion Analysis. Microchem. J. 2018, 140, 38–46. [Google Scholar] [CrossRef]
- Hong, Y.; Birse, N.; Quinn, B.; Li, Y.; Jia, W.; McCarron, P.; Wu, D.; Da Silva, G.R.; Vanhaecke, L.; Van Ruth, S.; et al. Data Fusion and Multivariate Analysis for Food Authenticity Analysis. Nat. Commun. 2023, 14, 3309. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhao, J.; Chen, Q.; Zhang, Y. Nondestructive Measurement of Total Volatile Basic Nitrogen (TVB-N) in Pork Meat by Integrating near Infrared Spectroscopy, Computer Vision and Electronic Nose Techniques. Food Chem. 2014, 145, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Lösel, H.; Brockelt, J.; Gärber, F.; Teipel, J.; Kuballa, T.; Seifert, S.; Fischer, M. Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites 2023, 13, 882. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-P.; Chen, P.; Dai, J.-W.; Liu, D.; Li, J.-Y.; Xu, Y.-P.; Chu, X.-L. Recent Advances of Chemometric Calibration Methods in Modern Spectroscopy: Algorithms, Strategy, and Related Issues. TrAC Trends Anal. Chem. 2022, 153, 116648. [Google Scholar] [CrossRef]
- Yao, S.; Li, J.; Duan, Z.; Li, T.; Wang, Y. Fusion of Ultraviolet and Infrared Spectra Using Support Vector Machine and Random Forest Models for the Discrimination of Wild and Cultivated Mushrooms. Anal. Lett. 2020, 53, 1019–1033. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Xia, J.; Xiong, Y.; Min, S. Quantitative Analysis of Honey Adulteration by Spectrum Analysis Combined with Several High-Level Data Fusion Strategies. Vib. Spectrosc. 2020, 108, 103060. [Google Scholar] [CrossRef]
- Liu, J.; Tang, H.; Zheng, G.C.; Li, J.; You, J. GAT and 3D-CNN Based Multi-Feature Fusion High-Spectrum Image Classification Method, Involves Performing Linear Feature Fusion to Advanced Feature Image and Super-Pixel-Level Feature Image, and Outputting Classification Result of Image Data to Be Tested. CN115375941-A.
- Ríos-Reina, R.; Azcarate, S.M.; Camiña, J.M.; Goicoechea, H.C. Multi-Level Data Fusion Strategies for Modeling Three-Way Electrophoresis Capillary and Fluorescence Arrays Enhancing Geographical and Grape Variety Classification of Wines. Anal. Chim. Acta 2020, 1126, 52–62. [Google Scholar] [CrossRef]
- Xian, G. Multiple Microphone Speaker Recognition System for Second Language Based on Biomimetic Pattern Recognition with Big Data Fusion. IOP Conf. Ser. Mater. Sci. Eng. 2020, 790, 012144. [Google Scholar] [CrossRef]
- Fang, J.; Hou, H.; Bi, Z.M.; Jin, D.; Han, L.; Yang, J.; Dai, S. Data Fusion in Forecasting Medical Demands Based on Spectrum of Post-Earthquake Diseases. J. Ind. Inf. Integr. 2021, 24, 100235. [Google Scholar] [CrossRef]
- Ihnaini, B.; Khan, M.A.; Khan, T.A.; Abbas, S.; Daoud, M.S.; Ahmad, M.; Khan, M.A. A Smart Healthcare Recommendation System for Multidisciplinary Diabetes Patients with Data Fusion Based on Deep Ensemble Learning. Comput. Intell. Neurosci. 2021, 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, R.; Wang, J.; Yu, T.; Su, A. Nonlinear State Estimation with Delayed Measurements Using Data Fusion Technique and Cubature Kalman Filter for Chemical Processes. Chem. Eng. Res. Des. 2019, 141, 502–515. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Steinier, J.; Termonia, Y.; Deltour, J. Smoothing and Differentiation of Data by Simplified Least Square Procedure. Anal. Chem. 1972, 44, 1906–1909. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing Methods for Near-infrared Spectrum Calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Brás, L.P.; Bernardino, S.A.; Lopes, J.A.; Menezes, J.C. Multiblock PLS as an Approach to Compare and Combine NIR and MIR Spectra in Calibrations of Soybean Flour. Chemom. Intell. Lab. Syst. 2005, 75, 91–99. [Google Scholar] [CrossRef]
- Pizarro, C.; Rodríguez-Tecedor, S.; Pérez-del-Notario, N.; Esteban-Díez, I.; González-Sáiz, J.M. Classification of Spanish Extra Virgin Olive Oils by Data Fusion of Visible Spectroscopic Fingerprints and Chemical Descriptors. Food Chem. 2013, 138, 915–922. [Google Scholar] [CrossRef]
- Casale, M.; Oliveri, P.; Casolino, C.; Sinelli, N.; Zunin, P.; Armanino, C.; Forina, M.; Lanteri, S. Characterisation of PDO Olive Oil Chianti Classico by Non-Selective (UV–Visible, NIR and MIR Spectroscopy) and Selective (Fatty Acid Composition) Analytical Techniques. Anal. Chim. Acta 2012, 712, 56–63. [Google Scholar] [CrossRef]
- Vera, L.; Aceña, L.; Guasch, J.; Boqué, R.; Mestres, M.; Busto, O. Discrimination and Sensory Description of Beers through Data Fusion. Talanta 2011, 87, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.-X.; Li, X.-J.; Zhang, H.-Y. Where Is the Hope for Drug Discovery? Let History Tell the Future. Drug Discov. Today 2009, 14, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shen, Y.; Yao, C.; Guo, D. Quality Assessment of Herbal Medicines Based on Chemical Fingerprints Combined with Chemometrics Approach: A Review. J. Pharm. Biomed. Anal. 2020, 185, 113215. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Chhabra, V.; Rawal, R.K.; Sharma, S. Chemometrics: A New Scenario in Herbal Drug Standardization. J. Pharm. Anal. 2014, 4, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiao, C.; Xu, Z.; Wu, Y.; Forsberg, E.; Peng, X.; He, S. Determination of Geographic Origins and Types of Lindera Aggregata Samples Using a Portable Short-Wave Infrared Hyperspectral Imager. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 279, 121370. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liang, Y.-Z.; Liu, H. Chemometrics Applied to Quality Control and Metabolomics for Traditional Chinese Medicines. J. Chromatogr. B 2016, 1015–1016, 82–91. [Google Scholar] [CrossRef]
- Yu, M.; Xu, G.; Qin, M.; Li, Y.; Guo, Y.; Ma, Q. Multiple Fingerprints and Spectrum-Effect Relationship of Polysaccharides from Saposhnikoviae Radix. Molecules 2022, 27, 5278. [Google Scholar] [CrossRef]
- Li, X.; Gong, D.; Sun, G.; Zhang, H.; Sun, W. High Performance Liquid Chromatography Three-Wavelength Fusion Fingerprint Combined with Electrochemical Fingerprint and Antioxidant Method to Evaluate the Quality Consistency of Mingmu Dihuang Pill. J. Chromatogr. A 2022, 1681, 463448. [Google Scholar] [CrossRef]
- Liang, Y.-Z.; Xie, P.-S.; Chan, K. Chromatographic Fingerprinting and Metabolomics for Quality Control of TCM. Comb Chem. High Throughput Screen. 2010, 13, 943–953. [Google Scholar] [CrossRef]
- Kabir, M.H.; Guindo, M.L.; Chen, R.; Liu, F.; Luo, X.; Kong, W. Deep Learning Combined with Hyperspectral Imaging Technology for Variety Discrimination of Fritillaria Thunbergii. Molecules 2022, 27, 6042. [Google Scholar] [CrossRef]
- Jiao, C.; Xu, Z.; Bian, Q.; Forsberg, E.; Tan, Q.; Peng, X.; He, S. Machine Learning Classification of Origins and Varieties of Tetrastigma Hemsleyanum Using a Dual-Mode Microscopic Hyperspectral Imager. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120054. [Google Scholar] [CrossRef] [PubMed]
- Ai, K.; Yuan, D.; Zheng, J. Experimental Research on the Antitumor Effect of Human Gastric Cancer Cells Transplanted in Nude Mice Based on Deep Learning Combined with Spleen-Invigorating Chinese Medicine. Comput. Math. Methods Med. 2022, 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Chen, Y.; Gao, W.; Liu, J.; Huang, Q.; Yan, C.; Huang, X.; Ding, C. An Improved Deep Learning Model: S-TextBLCNN for Traditional Chinese Medicine Formula Classification. Front. Genet. 2021, 12, 807825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, L.; Song, M.; Yan, J.; Shi, J.; Yao, Y. Evaluating the Traditional Chinese Medicine (TCM) Officially Recommended in China for COVID-19 Using Ontology-Based Side-Effect Prediction Framework (OSPF) and Deep Learning. J. Ethnopharmacol. 2021, 272, 113957. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-F.; Li, W.-X.; Zhang, F.; Li, Y.-H.; Cao, Y.-J.; Zhao, Y.; Li, X.-L.; Ma, Z.-J. Discrimination of Radix Polygoni Multiflori from Different Geographical Areas by UPLC-QTOF/MS Combined with Chemometrics. Chin. Med. 2017, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Chen, Y.; Wu, H.-L.; Chen, Y.; Wang, T.; Yang, J.; Fu, H.-Y.; Yang, X.-L.; Li, X.-F.; Yu, R.-Q. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics to Identify the Origin of Chinese Medicinal Materials. RSC Adv. 2022, 12, 16886–16892. [Google Scholar] [CrossRef]
- Tao, L.; Liu, B.; Jin, Y.; Sun, D.; Liu, X.; Chen, Y.; Wu, Y. Characterization of Toad Skin for Traditional Chinese Medicine by Near-Infrared Spectroscopy and Chemometrics. Anal. Lett. 2017, 50, 1292–1306. [Google Scholar] [CrossRef]
- Wang, N.; Li, Z.-Y.; Zheng, X.-L.; Li, Q.; Yang, X.; Xu, H. Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker. Molecules 2018, 23, 856. [Google Scholar] [CrossRef]
- Kellogg, J.J.; Wallace, E.D.; Graf, T.N.; Oberlies, N.H.; Cech, N.B. Conventional and Accelerated-Solvent Extractions of Green Tea (Camellia Sinensis) for Metabolomics-Based Chemometrics. J. Pharm. Biomed. Anal. 2017, 145, 604–610. [Google Scholar] [CrossRef]
- Liu, R.; Sun, Q.; Hu, T.; Li, L.; Nie, L.; Wang, J.; Zhou, W.; Zang, H. Multi-Parameters Monitoring during Traditional Chinese Medicine Concentration Process with near Infrared Spectroscopy and Chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 192, 75–81. [Google Scholar] [CrossRef]
- Han, J.; Xu, K.; Yan, Q.; Sui, W.; Zhang, H.; Wang, S.; Zhang, Z.; Wei, Z.; Han, F. Qualitative and Quantitative Evaluation of Flos Puerariae by Using Chemical Fingerprint in Combination with Chemometrics Method. J. Pharm. Anal. 2022, 12, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, W. Metabolomics: An Integral Technique in Systems Biology. Bioanalysis 2010, 2, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Dong, N.; Liu, S.; Yi, Z.; Zhang, Y. Chemical Features of Pericarpium Citri Reticulatae and Pericarpium Citri Reticulatae Viride Revealed by GC–MS Metabolomics Analysis. Food Chem. 2015, 186, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Charilaou, P.; Battat, R. Machine Learning Models and Over-Fitting Considerations. World J Gastroenterol. 2022, 28, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Casian, T.; Nagy, B.; Kovács, B.; Galata, D.L.; Hirsch, E.; Farkas, A. Challenges and Opportunities of Implementing Data Fusion in Process Analytical Technology—A Review. Molecules 2022, 27, 4846. [Google Scholar] [CrossRef] [PubMed]
- Belkin, M.; Hsu, D.; Ma, S.; Mandal, S. Reconciling Modern Machine-Learning Practice and the Classical Bias–Variance Trade-Off. Proc. Natl. Acad. Sci. USA 2019, 116, 15849–15854. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, L.; You, T.; Lu, A. Measurement of Potentially Toxic Elements in the Soil through NIR, MIR, and XRF Spectral Data Fusion. Comput. Electron. Agric. 2021, 187, 106257. [Google Scholar] [CrossRef]
- Fu, H.; Shi, Q.; Wei, L.; Xu, L.; Guo, X.; Hu, O.; Lan, W.; Xie, S.; Yang, T. Rapid Recognition of Geoherbalism and Authenticity of a Chinese Herb by Data Fusion of Near-Infrared Spectroscopy (NIR) and Mid-Infrared (MIR) Spectroscopy Combined with Chemometrics. J. Spectrosc. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Pei, Y.; Zuo, Z.; Zhang, Q.; Wang, Y. Data Fusion of Fourier Transform Mid-Infrared (MIR) and Near-Infrared (NIR) Spectroscopies to Identify Geographical Origin of Wild Paris Polyphylla Var. Yunnanensis. Molecules 2019, 24, 2559. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, X.; Zhang, Z.; Zhu, R. Data Fusion of Near-Infrared and Mid-Infrared Spectra for Identification of Rhubarb. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 72–79. [Google Scholar] [CrossRef]
- Qi, L.; Ma, Y.; Zhong, F.; Shen, C. Comprehensive Quality Assessment for Rhizoma Coptidis Based on Quantitative and Qualitative Metabolic Profiles Using High Performance Liquid Chromatography, Fourier Transform near-Infrared and Fourier Transform Mid-Infrared Combined with Multivariate Statistical Analysis. J. Pharm. Biomed. Anal. 2018, 161, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, X.; Li, L.; Li, H.; Gao, L.; Yuan, X.; Du, H.; Guan, Y.; Zang, H. Multi Critical Quality Attributes Monitoring of Chinese Oral Liquid Extraction Process with a Spectral Sensor Fusion Strategy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 278, 121317. [Google Scholar] [CrossRef] [PubMed]
- Hai, C.; Long, W.; Suo, Y.; Lu, H.; Chen, H.; Yang, X.; Yang, J.; Fu, H. Nano-Effect Multivariate Fusion Spectroscopy Combined with Chemometrics for Accurate Identification the Cultivation Methods and Growth Years of Dendrobium Huoshanense. Microchem. J. 2022, 179, 107556. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Tang, L.; Li, L.; Zhou, Q.; Li, Y.-J.; Li, J.; Wang, Y.-Z. Geographic Authentication of Eucommia Ulmoides Leaves Using Multivariate Analysis and Preliminary Study on the Compositional Response to Environment. Front. Plant Sci. 2020, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.-Y.; Wang, Y.-Z. FT-MIR and NIR Spectral Data Fusion: A Synergetic Strategy for the Geographical Traceability of Panax Notoginseng. Anal. Bioanal. Chem. 2018, 410, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Song, J.; Peng, L.; Gao, L.; Liu, X.; Xie, L.; Li, G. Improving Identification Ability of Adulterants in Powdered Panax Notoginseng Using Particle Swarm Optimization and Data Fusion. Infrared Phys. Technol. 2019, 103, 103101. [Google Scholar] [CrossRef]
- Li, M.-X.; Fang, H.; Chen, Y.; Wang, T.; Yang, J.; Fu, H.-Y.; Yang, X.-L.; Li, X.-F.; Chen, Z.-P.; Yu, R.-Q. Data Fusion of Synchronous Fluorescence and Surface Enhanced Raman Scattering Spectroscopies for Geographical Origin Traceability of Atractylodes Macrocephala Koidz. Spectrosc. Lett. 2022, 55, 290–301. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.-G.; Feng, T.; Zhang, T.-L.; Feng, Y.-Q.; Li, H. Discrimination of Radix Astragali According to Geographical Regions by Data Fusion of Laser Induced Breakdown Spectroscopy (LIBS) and Infrared Spectroscopy (IR) Combined with Random Forest (RF). Chin. J. Anal. Chem. 2022, 50, 100057. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, Y.; Qu, H. Data Fusion Strategy Based on near Infrared Spectra and Ultraviolet Spectra for Simultaneous Determination of Ginsenosides and Saccharides in Chinese Herbal Injection. Anal. Methods 2013, 5, 4467. [Google Scholar] [CrossRef]
- Zhang, J.S.; Wang, Y.M.; Song, X.H.; Sun, L.X.; Sun, Y.Z. A Comparison of Mineral Element Contents Among Different Types of Ginseng Using Inductively Coupled Plasma-Atomic Emission Spectroscopy. Microchem. J. 1994, 50, 131–135. [Google Scholar] [CrossRef]
- Yan, B.; Sun, G. Monitoring Quality Consistency of Liuwei Dihuang Pill by Integrating the Ultraviolet Spectroscopic Fingerprint, a Multi-Wavelength Fusion Fingerprint Method, and Antioxidant Activities. J. Sep. Sci. 2018, 41, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Ye-xia, Y.; Li, L.; Yuan-zhong, W. Study on Differentiation of Swertia Leducii and Its Closely Relative Species Based on Data Fusion of Spectra and Chromatography. Spectrosc. Spectr. Anal. 2020, 40, 2440–2446. [Google Scholar] [CrossRef]
- Wu, X.-M.; Zuo, Z.-T.; Zhang, Q.-Z.; Wang, Y.-Z. Classification of Paris Species According to Botanical and Geographical Origins Based on Spectroscopic, Chromatographic, Conventional Chemometric Analysis and Data Fusion Strategy. Microchem. J. 2018, 143, 367–378. [Google Scholar] [CrossRef]
- Wingfield, J.; Wilson, I.D. Advances in Mass Spectrometry Within Drug Discovery. SLAS Discov. 2016, 21, 109–110. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Lin, Z.; Xu, B.; Wang, Y.; Shi, X.; Qiao, Y.; Zhang, J. Metabolomics Data Fusion between near Infrared Spectroscopy and High-Resolution Mass Spectrometry: A Synergetic Approach to Boost Performance or Induce Confusion. Talanta 2018, 189, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-C.; Canellas, E.; Asensio, E.; Nerín, C. Predicting the Antioxidant Capacity and Total Phenolic Content of Bearberry Leaves by Data Fusion of UV–Vis Spectroscopy and UHPLC/Q-TOF-MS. Talanta 2020, 213, 120831. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zhang, J.; Zhao, Y.; Zuo, Z.; Wang, Y.-Z.; Jin, H. Characterization of Gentiana Rigescen s by Ultraviolet–Visible and Infrared Spectroscopies with Chemometrics. Anal. Lett. 2017, 50, 1497–1511. [Google Scholar] [CrossRef]
- Yao, S.; Li, T.; Liu, H.-G.; Li, J.-Q.; Wang, Y.-Z. Geographic Characterization of Leccinum Rugosiceps by Ultraviolet and Infrared Spectral Fusion. Anal. Lett. 2017, 50, 2257–2269. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.; Huang, H.; Wang, Y. Comparison and Quantitative Analysis of Wild and Cultivated Macrohyporia Cocos Using Attenuated Total Refection-Fourier Transform Infrared Spectroscopy Combined with Ultra-Fast Liquid Chromatography. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117633. [Google Scholar] [CrossRef]
- Li, L.; Zhang, S.; Zuo, Z.; Wang, Y. Data Fusion of Multiple-information Strategy Based on Fourier Transform near Infrared Spectroscopy and Fourier-transform Mid Infrared for Geographical Traceability of Wolfiporia Cocos Combined with Chemometrics. J. Chemom. 2022, 36, 9. [Google Scholar] [CrossRef]
- Wu, X.-M.; Zhang, Q.-Z.; Wang, Y.-Z. Traceability of Wild Paris Polyphylla Smith Var. Yunnanensis Based on Data Fusion Strategy of FT-MIR and UV–Vis Combined with SVM and Random Forest. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 205, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.Z.; Yang, M.Q.; Yang, W.Z.; Yang, S.B.; Zhang, J.Y. Identification and Evaluation of Polygonatum Kingianum with Different Growth Ages Based on Data Fusion Strategy. Microchem. J. 2021, 160, 105662. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Sun, H.; Dai, J.; Zhao, M.; Teng, C.; Ke, Z.; Yang, M.; Zhong, L.; Zhu, W. Application of a Data Fusion Strategy Combined with Multivariate Statistical Analysis for Quantification of Puerarin in Radix Puerariae. Vib. Spectrosc. 2020, 108, 103057. [Google Scholar] [CrossRef]
- Tao, L.; Via, B.; Wu, Y.; Xiao, W.; Liu, X. NIR and MIR Spectral Data Fusion for Rapid Detection of Lonicera Japonica and Artemisia Annua by Liquid Extraction Process. Vib. Spectrosc. 2019, 102, 31–38. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, H.-Y.; Wang, Y.-Z. Authentication of Dendrobium Officinale from Similar Species with Infrared and Ultraviolet-Visible Spectroscopies with Data Visualization and Mining. Anal. Lett. 2020, 53, 1774–1793. [Google Scholar] [CrossRef]
- Tian, M.; Ma, X.; Liang, M.; Zang, H. Application of Rapid Identification and Determination of Moisture Content of Coptidis Rhizoma from Different Species Based on Data Fusion. J. AOAC Int. 2023, 106, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Bai, X.; Gao, P.; He, Y. Application of Convolutional Neural Network-Based Feature Extraction and Data Fusion for Geographical Origin Identification of Radix Astragali by Visible/Short-Wave Near-Infrared and Near Infrared Hyperspectral Imaging. Sensors 2020, 20, 4940. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Li, J.; Wang, Y. Solving the Identification Problems of Bolete Origins Based on Multiple Data Processing: Take Boletus Bainiugan as an Example. J. Food Compos. Anal. 2023, 124, 105693. [Google Scholar] [CrossRef]
- Masnan, M.J.; Mahat, N.I.; Zakaria, A.; Shakaff, A.Y.M.; Adom, A.H.; Sa’ad, F.S.A. Enhancing Classification Performance of Multisensory Data through Extraction and Selection of Features. Procedia Chem. 2012, 6, 132–140. [Google Scholar] [CrossRef]
- Wang, Q.-Q.; Huang, H.-Y.; Wang, Y.-Z. Geographical Authentication of Macrohyporia Cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy. Molecules 2019, 24, 1320. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.; Wang, Y.; Wang, Y. Comparison of Geographical Traceability of Wild and Cultivated Macrohyporia Cocos with Different Data Fusion Approaches. J. Anal. Methods Chem. 2021, 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lan, Z.; Zhang, Y.; Sun, Y.; Ji, D.; Wang, S.; Lu, T.; Cao, H.; Meng, J. A Mid-Level Data Fusion Approach for Evaluating the Internal and External Changes Determined by FT-NIR, Electronic Nose and Colorimeter in Curcumae Rhizoma Processing. J. Pharm. Biomed. Anal. 2020, 188, 113387. [Google Scholar] [CrossRef] [PubMed]
- Massaro, A.; Negro, A.; Bragolusi, M.; Miano, B.; Tata, A.; Suman, M.; Piro, R. Oregano Authentication by Mid-Level Data Fusion of Chemical Fingerprint Signatures Acquired by Ambient Mass Spectrometry. Food Control. 2021, 126, 108058. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Wang, L.; Shi, Z.-Q.; Li, P.; Li, H.-J. A Metabolomic Strategy Based on Integrating Headspace Gas Chromatography-Mass Spectrometry and Liquid Chromatography-Mass Spectrometry to Differentiate the Five Cultivars of Chrysanthemum Flower. RSC Adv. 2018, 8, 9074–9082. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Razmovski-Naumovski, V.; Li, K.M.; Li, G.Q.; Chan, K. Differentiating Puerariae Lobatae Radix and Puerariae Thomsonii Radix Using HPTLC Coupled with Multivariate Classification Analyses. J. Pharm. Biomed. Anal. 2014, 95, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gong, D.; Liu, X.; Sun, G.; Sun, W. Quality and Antioxidant Activity Evaluation of Dandelion by HPLC with Five-Wavelength Fusion Fingerprint. New J. Chem. 2021, 45, 9856–9863. [Google Scholar] [CrossRef]
- Shen, T.; Yu, H.; Wang, Y. Assessing Geographical Origin of Gentiana Rigescens Using Untargeted Chromatographic Fingerprint, Data Fusion and Chemometrics. Molecules 2019, 24, 2562. [Google Scholar] [CrossRef]
- Dubin, E.; Spiteri, M.; Dumas, A.-S.; Ginet, J.; Lees, M.; Rutledge, D.N. Common Components and Specific Weights Analysis: A Tool for Metabolomic Data Pre-Processing. Chemom. Intell. Lab. Syst. 2016, 150, 41–50. [Google Scholar] [CrossRef]
- Smith, R.; Ventura, D.; Prince, J.T. LC-MS Alignment in Theory and Practice: A Comprehensive Algorithmic Review. Brief. Bioinform. 2015, 16, 104–117. [Google Scholar] [CrossRef]
- Charve, J.; Chen, C.; Hegeman, A.D.; Reineccius, G.A. Evaluation of Instrumental Methods for the Untargeted Analysis of Chemical Stimuli of Orange Juice Flavour: Evaluation of Instrumental Methods for Flavoromic Research. Flavour Fragr. J. 2011, 26, 429–440. [Google Scholar] [CrossRef]
- Belmonte-Sánchez, J.R.; Romero-González, R.; Martínez Vidal, J.L.; Arrebola, F.J.; Garrido Frenich, A. 1H NMR and Multi-Technique Data Fusion as Metabolomic Tool for the Classification of Golden Rums by Multivariate Statistical Analysis. Food Chem. 2020, 317, 126363. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of Electronic Nose, Electronic Tongue and Computer Vision for Animal Source Food Authentication and Quality Assessment—A Review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, Y.; Zhan, W.; Zhu, D. Improved Thermal Infrared Image Super-Resolution Reconstruction Method Base on Multimodal Sensor Fusion. Entropy 2023, 25, 914. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Lin, Y.-B.; Hung, H.-N. CalibrationTalk: A Farming Sensor Failure Detection and Calibration Technique. IEEE Internet Things J. 2021, 8, 6893–6903. [Google Scholar] [CrossRef]
- Guerrero-Ibáñez, J.; Zeadally, S.; Contreras-Castillo, J. Sensor Technologies for Intelligent Transportation Systems. Sensors 2018, 18, 1212. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Li, Y.; Sun, J.; Zhou, D.; Zhang, Q. Multi-Sensor Fusion for Body Sensor Network in Medical Human–Robot Interaction Scenario. Inf. Fusion 2020, 57, 15–26. [Google Scholar] [CrossRef]
- Pan, M.H.; Su, Y.F.; Liu, X.J.; Xiang, S.Q.; Ding, Y.X.; Li, Q. Identification of Forsythia Suspensa (Thunb.) Vahl in Different Harvest Periods Using Intelligent Sensory Technologies, HPLC Characteristic Fingerprint Coupled with Chemometrics. Phytochem. Anal. 2022, 33, 490–501. [Google Scholar] [CrossRef]
- Fei, C.; Ren, C.; Wang, Y.; Li, L.; Li, W.; Yin, F.; Lu, T.; Yin, W. Identification of the Raw and Processed Crataegi Fructus Based on the Electronic Nose Coupled with Chemometric Methods. Sci. Rep. 2021, 11, 1849. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Lin, L.; Du, X.; Tang, S.; Liu, H.; Yang, H. The Qualitative and Quantitative Assessment of Xiaochaihu Granules Based on E-Eye, e-Nose, e-Tongue and Chemometrics. J. Pharm. Biomed. Anal. 2021, 205, 114298. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Z.; Zhang, B.; Du, J.; Li, W.; Wang, Z. Data Fusion of Electronic Noses and Electronic Tongues Aids in Botanical Origin Identification on Imbalanced Codonopsis Radix Samples. Sci. Rep. 2022, 12, 19120. [Google Scholar] [CrossRef]
- Miao, J.; Luo, Z.; Wang, Y.; Li, G. Comparison and Data Fusion of an Electronic Nose and Near-Infrared Reflectance Spectroscopy for the Discrimination of Ginsengs. Anal. Methods 2016, 8, 1265–1273. [Google Scholar] [CrossRef]
- Jing, W.; Zhao, X.; Li, M.; Hu, X.; Cheng, X.; Ma, S.; Wei, F. Application of Multiple-Source Data Fusion for the Discrimination of Two Botanical Origins of Magnolia Officinalis Cortex Based on E-Nose Measurements, E-Tongue Measurements, and Chemical Analysis. Molecules 2022, 27, 3892. [Google Scholar] [CrossRef] [PubMed]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data Fusion Methodologies for Food and Beverage Authentication and Quality Assessment—A Review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, R.; Ma, S.; Dai, S.; Zheng, J. Application of Data Fusion in Traditional Chinese Medicine: A Review. Sensors 2024, 24, 106. https://doi.org/10.3390/s24010106
Huang R, Ma S, Dai S, Zheng J. Application of Data Fusion in Traditional Chinese Medicine: A Review. Sensors. 2024; 24(1):106. https://doi.org/10.3390/s24010106
Chicago/Turabian StyleHuang, Rui, Shuangcheng Ma, Shengyun Dai, and Jian Zheng. 2024. "Application of Data Fusion in Traditional Chinese Medicine: A Review" Sensors 24, no. 1: 106. https://doi.org/10.3390/s24010106
APA StyleHuang, R., Ma, S., Dai, S., & Zheng, J. (2024). Application of Data Fusion in Traditional Chinese Medicine: A Review. Sensors, 24(1), 106. https://doi.org/10.3390/s24010106




