Development and Assessment of Multiple Illumination Color Fourier Ptychographic Microscopy for High Throughput Sample Digitization
Abstract
:1. Introduction
2. Materials and Methods
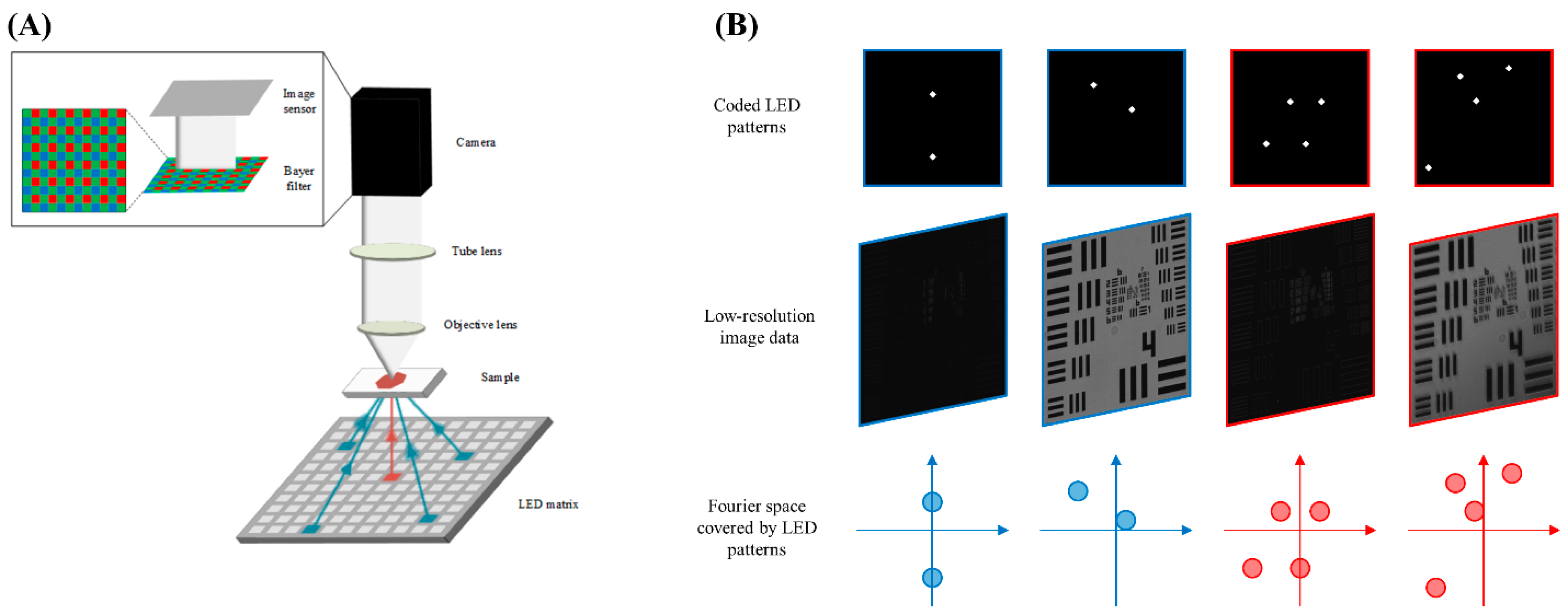
2.1. Multiplexed Fourier Ptychography
2.2. Experimental Setup
2.3. Multiplexed Illumination Pattern Design and FPM Image Reconstruction
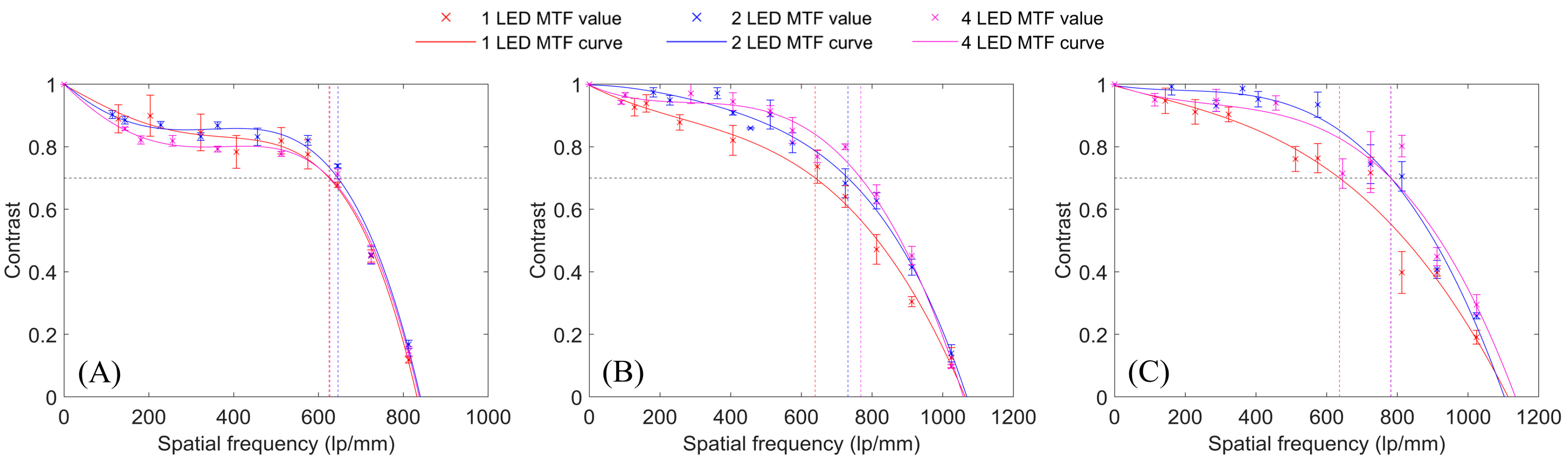
2.4. Multiplexed FPM System Performance Evaluation
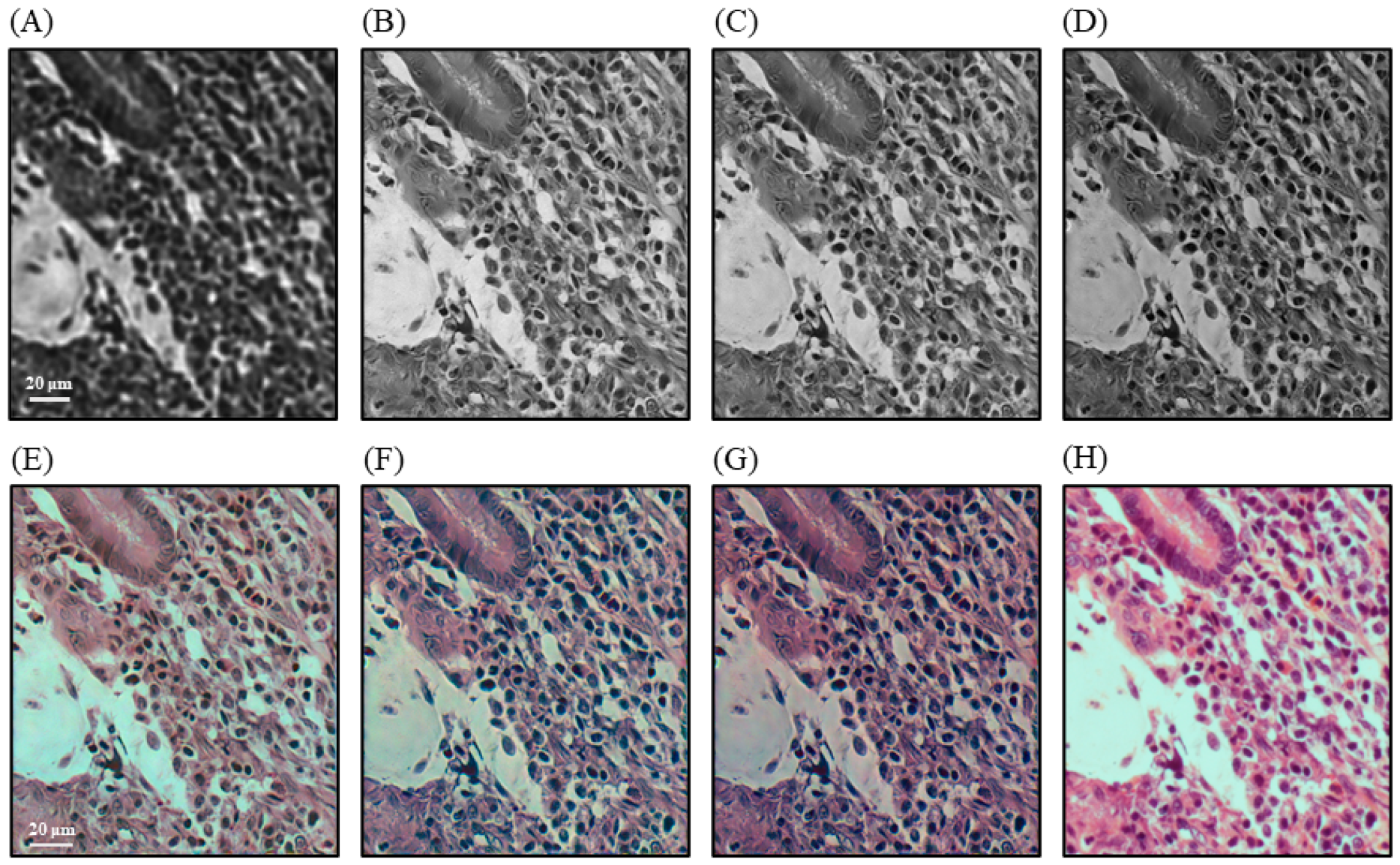
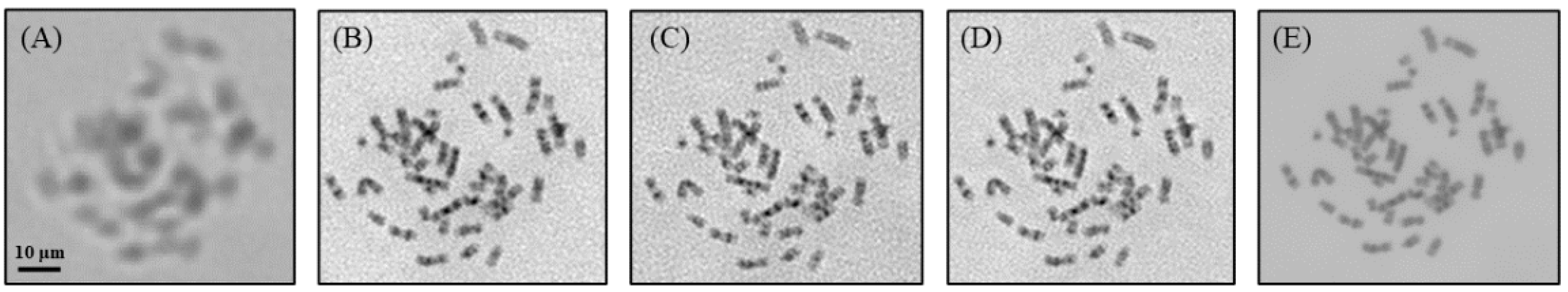
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, N.; Gupta, R.; Gupta, S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.; Kitsanta, P.; Perunovic, B.; Suvarna, S.K.; Bury, J. Digital pathology for intraoperative frozen section diagnosis of thoracic specimens: An evaluation of a system using remote sampling and whole slide imaging diagnosis. J. Clin. Pathol. 2020, 73, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Jahn, S.W.; Plass, M.; Moinfar, F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J. Clin. Med. 2020, 9, 3697. [Google Scholar] [CrossRef] [PubMed]
- Têtu, B.; Perron, É.; Louahlia, S.; Paré, G.; Trudel, M.C.; Meyer, J. The Eastern Québec Telepathology Network: A three-year experience of clinical diagnostic services. Diagn. Pathol. 2014, 9 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed]
- Sarode, V.R. The current practice of telecytology for rapid on-site evaluation (ROSE): Practical considerations and limitations. Semin. Diagn. Pathol. 2022, 39, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Tummidi, S.; Shankaralingappa, A.; Aravindakshan, R. Rapid on-site evaluation and cell blocks: Getting the most from the least invasive method in cytopathology. J. Am. Soc. Cytopathol. 2024, 13, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Adeniran, A.J. (Eds.) Rapid On-Site Evaluation (ROSE): A Practical Guide; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Cornish, T.C.; McClintock, D.S. Whole Slide Imaging and Telepathology. In Whole Slide Imaging: Current Applications and Future Directions; Parwani, A.V., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 117–152. [Google Scholar] [CrossRef]
- Zarella, M.D.; Rivera Alvarez, K. High-throughput whole-slide scanning to enable large-scale data repository building. J. Pathol. 2022, 257, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Horstmeyer, R.; Yang, C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat. Photonics 2013, 7, 739–745. [Google Scholar] [CrossRef]
- Hanna, M.G.; Parwani, A.; Sirintrapun, S.J. Whole Slide Imaging: Technology and Applications. Adv. Anat. Pathol. 2020, 27, 251–259. [Google Scholar] [CrossRef]
- Zheng, G.; Shen, C.; Jiang, S.; Song, P.; Yang, C. Concept, implementations and applications of Fourier ptychography. Nat. Rev. Phys. 2021, 3, 207–223. [Google Scholar] [CrossRef]
- Pan, A.; Zuo, C.; Yao, B. High-resolution and large field-of-view Fourier ptychographic microscopy and its applications in biomedicine. Rep. Prog. Progress. Phys. 2020, 83, 096101. [Google Scholar] [CrossRef]
- Zhang, K.; Lu, X.; Chen, X.; Zhang, R.; Fung, K.M.; Liu, H.; Zheng, B.; Li, S.; Qiu, Y. Using Fourier ptychography microscopy to achieve high-resolution chromosome imaging: An initial evaluation. J. Biomed. Opt. 2022, 27, 016504. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Zheng, G.; Yang, C. Embedded pupil function recovery for Fourier ptychographic microscopy. Opt. Express 2014, 22, 4960–4972. [Google Scholar] [CrossRef]
- Tian, L.; Li, X.; Ramchandran, K.; Waller, L. Multiplexed coded illumination for Fourier Ptychography with an LED array microscope. Biomed. Opt. Express 2014, 5, 2376–2389. [Google Scholar] [CrossRef] [PubMed]
- Claveau, R.; Manescu, P.; Elmi, M.; Pawar, V.; Shaw, M.; Fernandez-Reyes, D. Digital refocusing and extended depth of field reconstruction in Fourier ptychographic microscopy. Biomed. Opt. Express 2020, 11, 215–226. [Google Scholar] [CrossRef]
- Zhou, A.; Chen, N.; Wang, H.; Situ, G. Analysis of Fourier ptychographic microscopy with half of the captured images. J. Opt. 2018, 20, 095701. [Google Scholar] [CrossRef]
- Zhang, K.; Gilley, P.; Abdoli, N.; Chen, X.; Fung, K.-M.; Qiu, Y. Using symmetric illumination and color camera to achieve high throughput Fourier ptychographic microscopy. J. Biophotonics 2023, 16, e202200303. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, J.; Bian, Z.; Suo, J.; Zheng, G.; Dai, Q. Fourier ptychographic microscopy using wavelength multiplexing. J. Biomed. Opt. 2017, 22, 066006. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, D.; Liang, Y. Color Fourier ptychographic microscopy based on symmetrical illumination and wavelength multiplexing. J. Opt. 2020, 22, 065604. [Google Scholar] [CrossRef]
- Bian, L.; Suo, J.; Situ, G.; Zheng, G.; Chen, F.; Dai, Q. Content adaptive illumination for Fourier ptychography. Opt. Lett. 2014, 39, 6648–6651. [Google Scholar] [CrossRef]
- Guo, K.; Dong, S.; Nanda, P.; Zheng, G. Optimization of sampling pattern and the design of Fourier ptychographic illuminator. Opt. Express 2015, 23, 6171–6180. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Horstmeyer, R.; Zheng, G.; Yang, C. High numerical aperture Fourier ptychography: Principle, implementation and characterization. Opt. Express 2015, 23, 3472–3491. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Dong, S.; Zheng, G. Adaptive system correction for robust Fourier ptychographic imaging. Opt. Express 2013, 21, 32400–32410. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, Q.; Zhang, Y.; Zuo, C. Sampling criteria for Fourier ptychographic microscopy in object space and frequency space. Opt. Express 2016, 24, 15765–15781. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.J. Modern Optical Engineering: The Design of Optical Systems, 4th ed.; McGraw-Hill Education: New York, NY, USA, 2008. [Google Scholar]
- Qiu, Y.; Chen, X.; Li, Y.; Zheng, B.; Li, S.; Chen, W.R.; Liu, H. Impact of the optical depth of field on cytogenetic image quality. J. Biomed. Opt. 2012, 17, 096017. [Google Scholar] [CrossRef] [PubMed]
- Bradie, B. A Friendly Introduction to Numerical Analysis; Pearson Education: Hoboken, NJ, USA, 2006. [Google Scholar]
- Lam, E.Y.; Fung, G.S.K.; Lukac, R. Single-Sensor Imaging: Methods and Applications for Digital Cameras; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Tjio, J.H.; Levan, A. The Chromosome Number of Man. In Problems of Birth Defects: From Hippocrates to Thalidomide and After; Persaud, T.V.N., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 1977; pp. 112–118. [Google Scholar] [CrossRef]
- Yunis, J.J.; Sanchez, O. G-banding and chromosome structure. Chromosoma 1973, 44, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Vitkovski, T.; Bhuiya, T.; Esposito, M. Utility of telepathology as a consultation tool between an off-site surgical pathology suite and affiliated hospitals in the frozen section diagnosis of lung neoplasms. J. Pathol. Inform. 2015, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Aidukas, T.; Eckert, R.; Harvey, A.R.; Waller, L.; Konda, P.C. Low-cost, sub-micron resolution, wide-field computational microscopy using opensource hardware. Sci. Rep. 2019, 9, 7457. [Google Scholar] [CrossRef] [PubMed]
- Lin, O. Telecytology for Rapid On-Site Evaluation: Current Status. J. Am. Soc. Cytopathol. 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Sun, J.; Zuo, C.; Zhang, L.; Chen, Q. Resolution-enhanced Fourier ptychographic microscopy based on high-numerical-aperture illuminations. Sci. Rep. 2017, 7, 1187. [Google Scholar] [CrossRef]
- Nguyen, T.; Xue, Y.; Li, Y.; Tian, L.; Nehmetallah, G. Deep learning approach for Fourier ptychography microscopy. Opt. Express 2018, 26, 26470–26484. [Google Scholar] [CrossRef] [PubMed]
- Kellman, M.R.; Bostan, E.; Repina, N.A.; Waller, L. Physics-Based Learned Design: Optimized Coded-Illumination for Quantitative Phase Imaging. IEEE Trans. Comput. Imaging 2019, 5, 344–353. [Google Scholar] [CrossRef]
- Muthumbi, A.; Chaware, A.; Kim, K.; Zhou, K.C.; Konda, P.C.; Chen, R.; Judkewitz, B.; Erdmann, A.; Kappes, B.; Horstmeyer, R. Learned sensing: Jointly optimized microscope hardware for accurate image classification. Biomed. Opt. Express 2019, 10, 6351–6369. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilley, P.; Zhang, K.; Abdoli, N.; Sadri, Y.; Adhikari, L.; Fung, K.-M.; Qiu, Y. Development and Assessment of Multiple Illumination Color Fourier Ptychographic Microscopy for High Throughput Sample Digitization. Sensors 2024, 24, 4505. https://doi.org/10.3390/s24144505
Gilley P, Zhang K, Abdoli N, Sadri Y, Adhikari L, Fung K-M, Qiu Y. Development and Assessment of Multiple Illumination Color Fourier Ptychographic Microscopy for High Throughput Sample Digitization. Sensors. 2024; 24(14):4505. https://doi.org/10.3390/s24144505
Chicago/Turabian StyleGilley, Patrik, Ke Zhang, Neman Abdoli, Youkabed Sadri, Laura Adhikari, Kar-Ming Fung, and Yuchen Qiu. 2024. "Development and Assessment of Multiple Illumination Color Fourier Ptychographic Microscopy for High Throughput Sample Digitization" Sensors 24, no. 14: 4505. https://doi.org/10.3390/s24144505




