Isolation and Characterization of Exosomes from Cancer Cells Using Antibody-Functionalized Paddle Screw-Type Devices and Detection of Exosomal miRNA Using Piezoelectric Biosensor
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagent and Materials
2.2. Design and Fabrication of the SAW Sensor
2.3. Sensing and Fluidic Blocks of the SAW Sensor
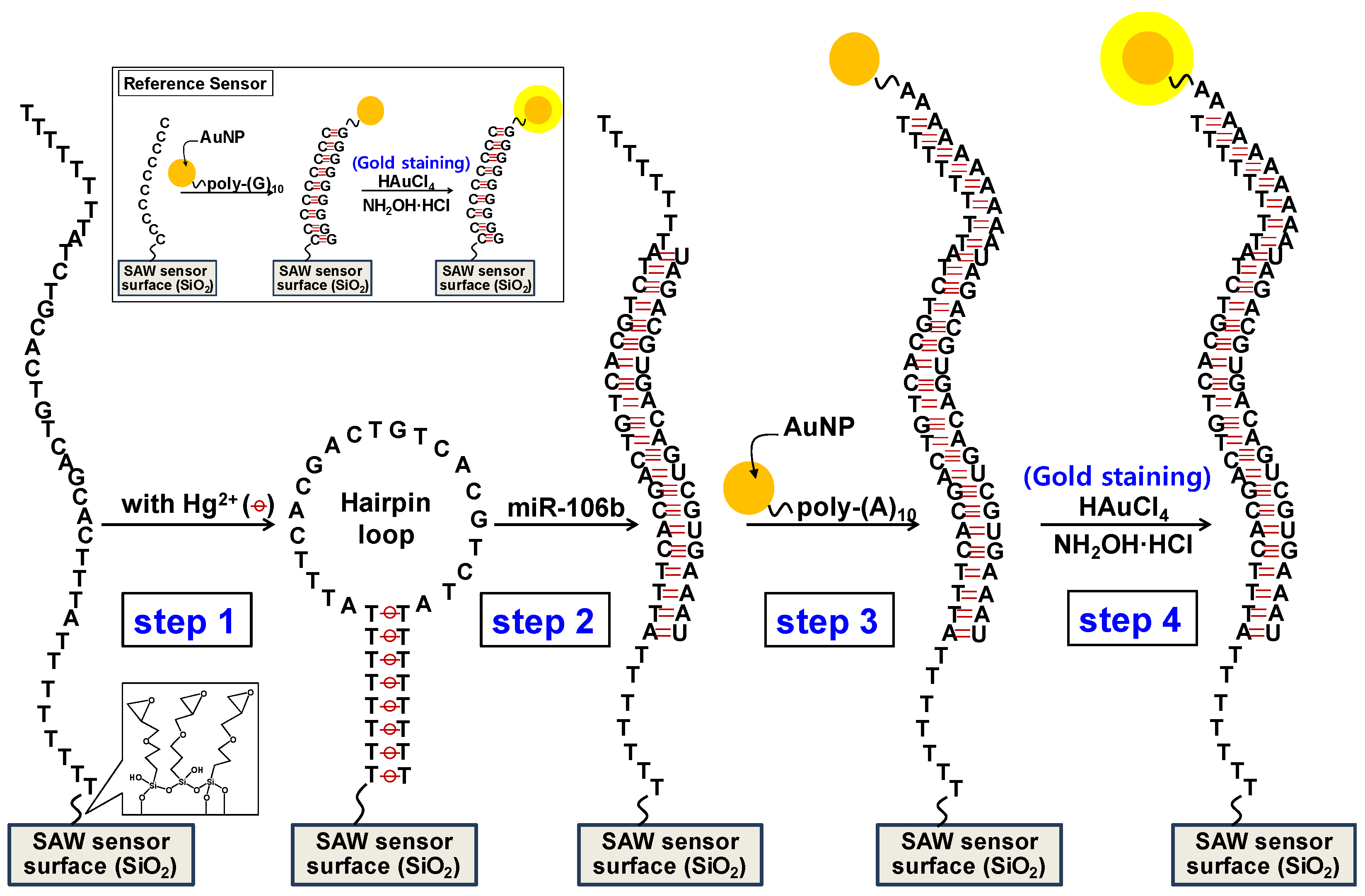
2.4. Detection of Synthetic miR-106b Using SAW Biosensor
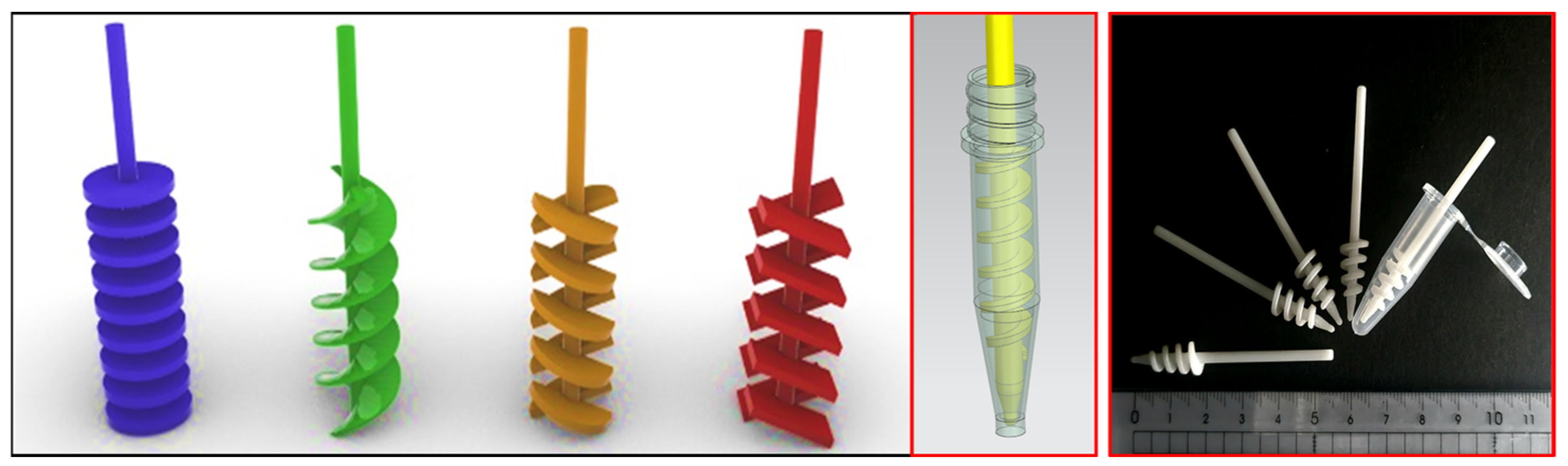
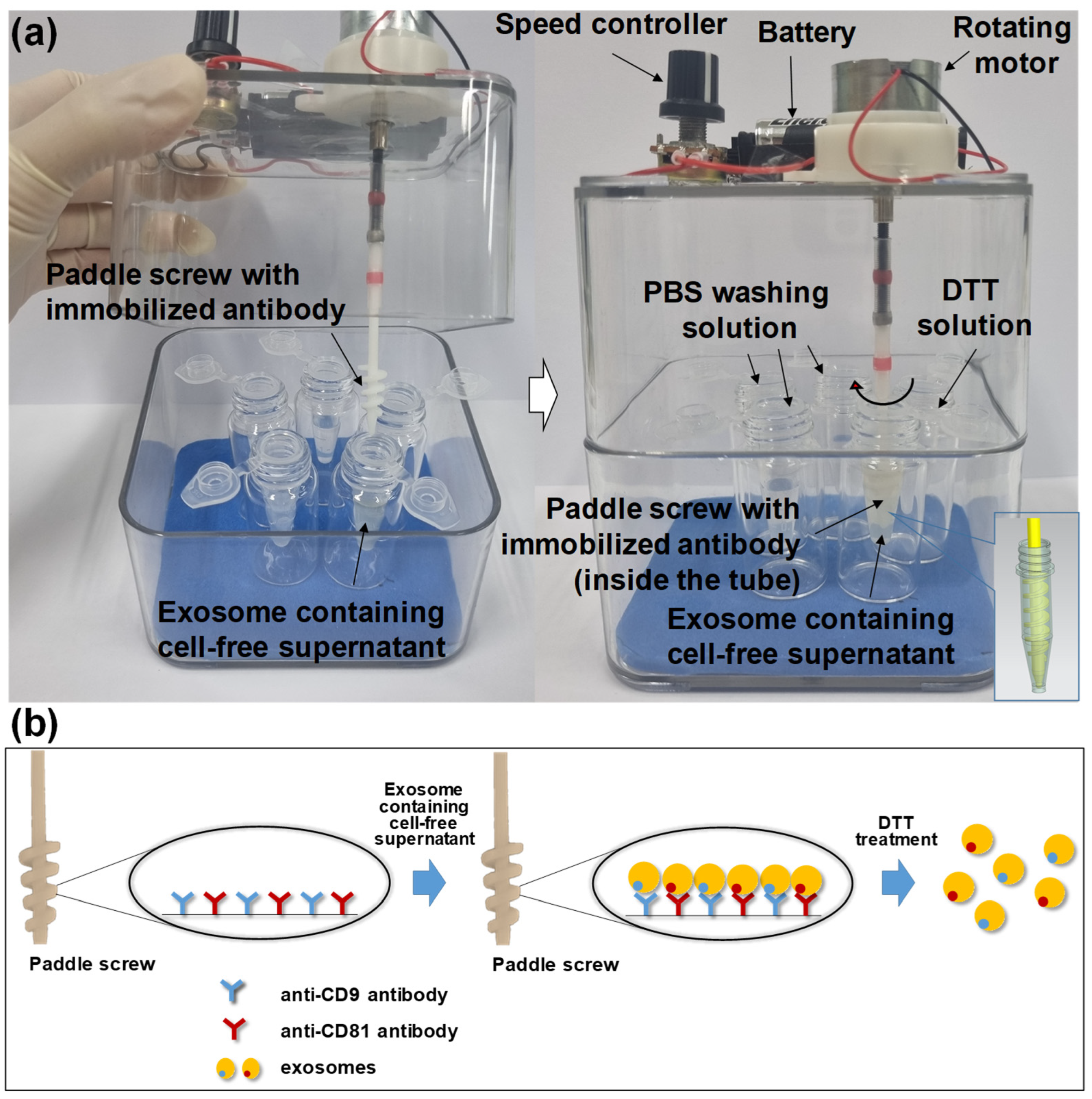
2.5. Preparation of Antibody-Conjugated Paddle Screw-Type Devices for Exosome Isolation
2.6. Isolation and Characterization of Cancer Cell-Derived Exosomes
2.7. RT-qPCR for Detecting miR-106b Using Total RNA Extracted from MCF-7 Human Breast Carcinoma Cell Line
3. Results and Discussion
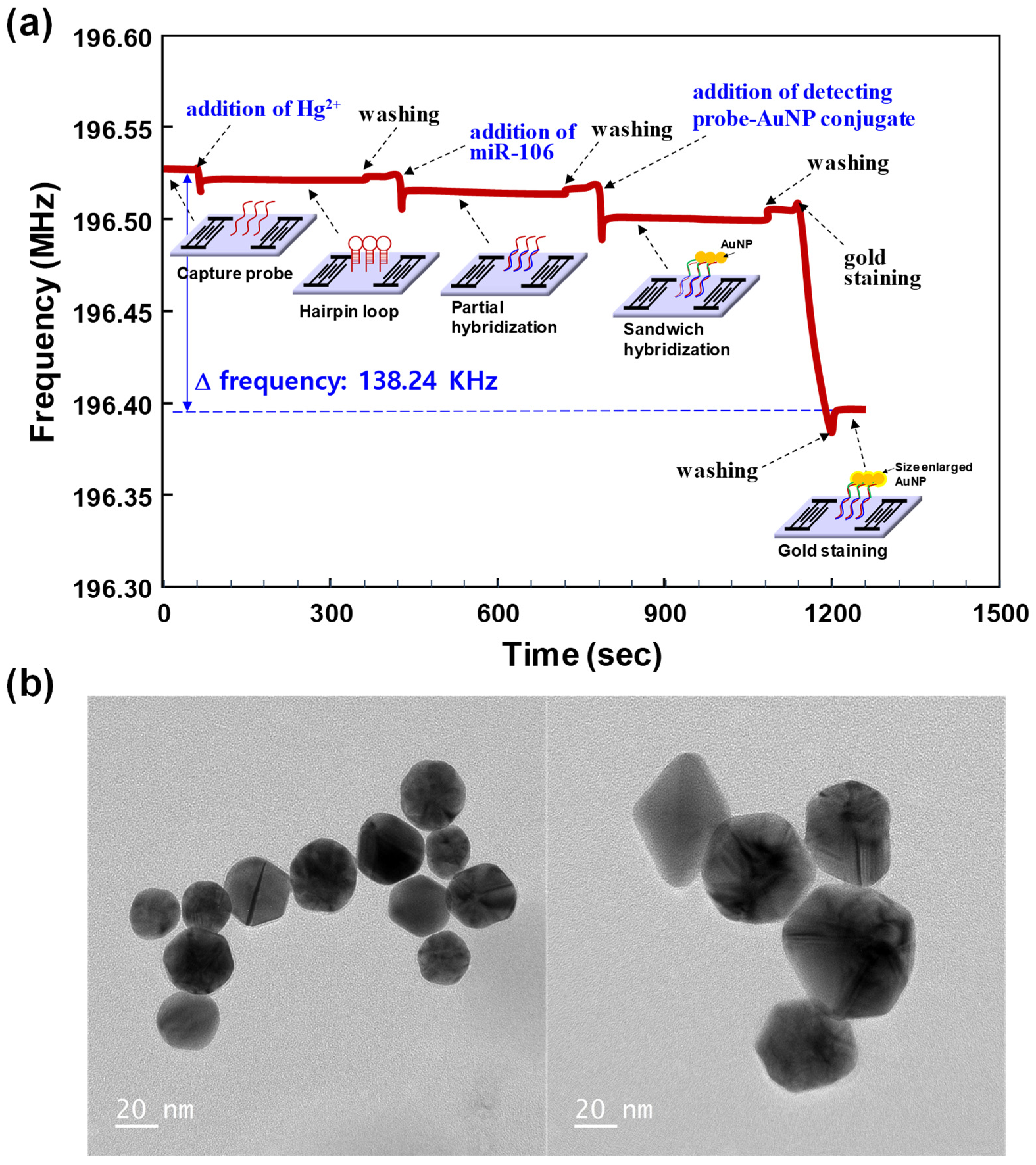
3.1. Sensing Principle of miR-106b Using the SAW Biosensor
3.2. Sensitivity and Selectivity of the SAW Biosensor for miR-106b Detection
3.3. Exosome Isolation from MCF-7 Cell Line and Its Characterization
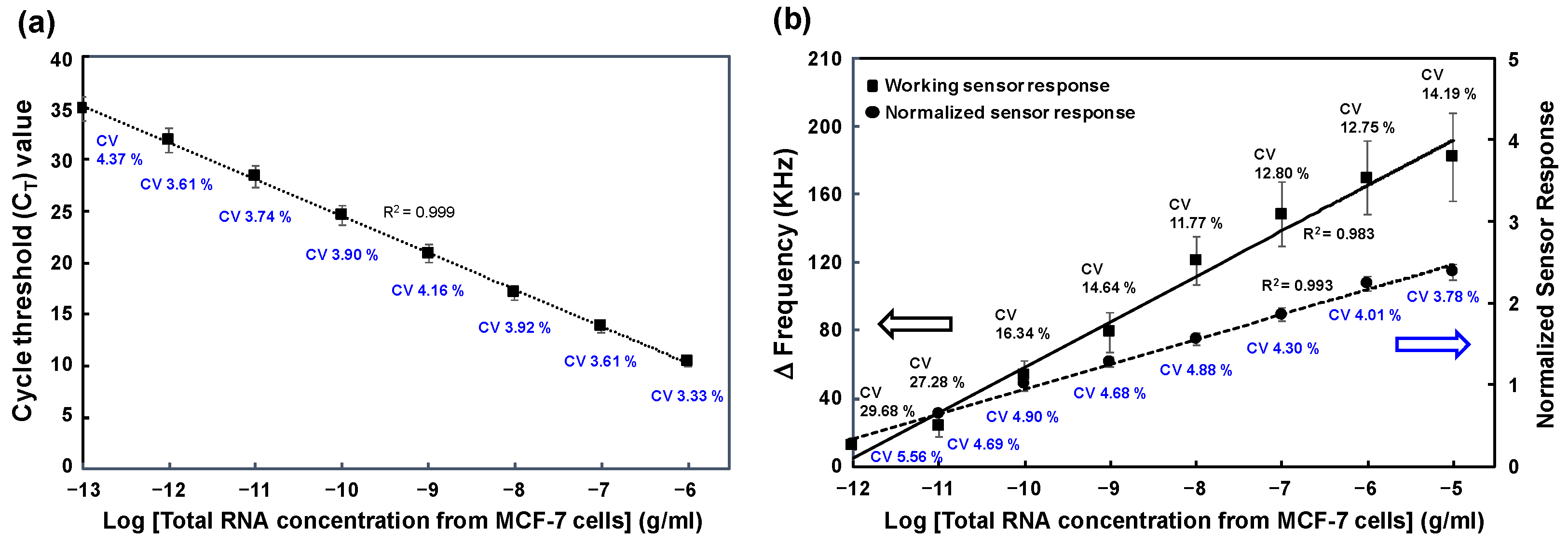
3.4. Comparison of miR-106b Detection in MCF-7 Cell-Derived RNA Samples Using RT-qPCR Assay and SAW Biosensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P.K. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, A.; Hu, J.; Feng, L.; Liu, L.; Shen, Z. Recent developments in isolating methods for exosomes. Front. Bioeng. Biotechnol. 2022, 10, 1100892. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on strategies and technologies for exosome isolation and purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Liu, W.-Z.; Ma, Z.-J.; Kang, X.-W. Current status and outlook of advances in exosome isolation. Anal. Bioanal. Chem. 2022, 414, 7123–7141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Lim, J.; Choi, M.; Lee, H.; Kim, Y.H.; Han, J.Y.; Lee, E.S.; Cho, Y. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J. Nanobiotechnol. 2019, 17, 1. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Ramirez, M.I.; Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.J.N. Technical challenges of working with extracellular vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef] [PubMed]
- Ciesla, M.; Skrzypek, K.; Kozakowska, M.; Loboda, A.; Jozkowicz, A.; Dulak, J. MicroRNAs as biomarkers of disease onset. Anal. Bioanal. Chem. 2011, 401, 2051–2061. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Speranza, M.C.; Frattini, V.; Pisati, F.; Kapetis, D.; Porrati, P. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget 2012, 3, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.K. miR-106b as an emerging therapeutic target in cancer. Genes. Dis. 2022, 9, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Sun, Z.; Wang, D.; Du, T. MiR-106b-5p regulates esophageal squamous cell carcinoma progression by binding to HPGD. BMC Cancer 2022, 22, 308. [Google Scholar] [CrossRef]
- Le, M.N.; Nguyen, T.A. Innovative microRNA quantification by qPCR. Mol. Ther. Nucleic Acids 2023, 31, 628–630. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kwak, J.; Lee, J.-H.; Lee, S.S. Efficient and accurate analysis of microRNA using a specific extension sequence. Mol. Biol. Rep. 2018, 45, 611–619. [Google Scholar] [CrossRef]
- Djebbi, K.; Shi, B.; Weng, T.; Bahri, M.; Elaguech, M.A.; Liu, L.; Tlili, C.; Wang, D. Highly sensitive fluorescence assay for miRNA detection: Investigation of the DNA spacer effect on the DSN enzyme activity toward magnetic-bead-tethered probes. ACS Omega 2022, 7, 2224–2234. [Google Scholar] [CrossRef]
- Yan, Y.; Hu, T.; Fanf, Y.; Xiang, X.; Ma, C. A fluorescence strategy for the rapid detection of miRNA-21 based on G-quadruplex and cyclic amplification signal. Anal. Biochem. 2022, 652, 114775. [Google Scholar] [CrossRef]
- Nemati, F.; Hosseini, M. Fluorescence turn-on detection of miRNA-155 based on hybrid Ce-MOF/PtNPs/graphene oxide serving as fluorescence quencher. J. Photochem. Photobiol. A Chem. 2022, 429, 113943. [Google Scholar] [CrossRef]
- Pothipor, C.; Aroomyadet, N.; Bamrungsap, S.; Jakmunee, J.; Ounnunkad, K. A highly sensitive electrochemical microRNA-21 biosensor based on intercalating methylene blue signal amplification and a highly dispersed gold nanoparticles/graphene/polypyrrole composite. Analyst 2021, 146, 2679–2688. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.L.; Oliveira, D.; Pego, A.P.; Santos, S.D.; Moreira, F.T.C. Electrochemical miRNA-34a-based biosensor for the diagnosis of Alzheimer’s disease. Bioelectrochemistry 2023, 154, 108553. [Google Scholar] [CrossRef]
- Cimmino, W.; Raucci, A.; Grosso, S.P.; Normanno, N.; Cinti, S. Enhancing sensitivity towards electrochemical miRNA detection using an affordable paper-based strategy. Anal. Bioanal. Chem. 2024, 416, 4227–4239. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, N.; Yang, A.; Xu, Z.; Zhang, W.; Liu, H.; Law, H.K. Ultrasensitive detection of ribonucleic acid biomarkers using portable sensing platforms based on organic electrochemical transistors. Anal. Chem. 2021, 93, 14359–14364. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; He, T.; Sun, Y.; Liu, Y.; Cao, Q.; Wang, Q.; Tang, H. An organic electrochemical transistor for determination of microRNA21 using gold nanoparticles and a capture DNA probe. Microchim. Acta 2018, 185, 408. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.S. QCM sensing of miR-21 by formation of microRNA–DNA hybrid duplexes and intercalation on surface-functionalized pyrene. Analyst 2019, 144, 6936–6943. [Google Scholar] [CrossRef]
- Li, K.; An, N.; Wu, L.; Wang, M.; Li, F.; Li, L. Absolute quantification of microRNAs based on mass transport limitation under a laminar flow SPR system. Biosens. Bioelectron. 2024, 244, 115776. [Google Scholar] [CrossRef]
- Wang, K.-H.; Chen, Y.-Y.; Wang, C.-H.; Hsu, H.-F.; Chau, L.-K.; Wang, S.-C.; Chen, Y.-L. Ultrasensitive amplification-free detection of circulating miRNA via droplet-based processing of SERS tag–miRNA–magnetic nanoparticle sandwich nanocomplexes on a paper-based electrowetting-on-dielectric platform. Analyst 2024, 149, 1981–1987. [Google Scholar] [CrossRef]
- Hu, Z.; Hu, Y.; Huang, L.; Zhong, W.; Zhang, J.; Lei, D.; Chen, Y.; Ni, Y.; Liu, Y. Recent Progress in Organic Electrochemical Transistor-Structured Biosensors. Biosensors 2024, 14, 330. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Das, P.K.; Bhethanabotla, V.R. Surface acoustic waves in biosensing applications. Sens. Actuators Rep. 2021, 3, 100041. [Google Scholar] [CrossRef]
- Han, S.B.; Lee, S.S. Simultaneous Detection of Exosomal microRNAs Isolated from Cancer Cells Using Surface Acoustic Wave Sensor Array with High Sensitivity and Reproducibility. Micromachines 2024, 15, 249. [Google Scholar] [CrossRef] [PubMed]
- Länge, K.; Rapp, B.E.; Rapp, M. Surface acoustic wave biosensors: A review. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [Google Scholar] [CrossRef]
- Baumgartner, K.; Westerhausen, C. Recent advances of surface acoustic wave-based sensors for noninvasive cell analysis. Biosens. Bioelectron. 2009, 24, 3120–3125. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-S.; Lee, J.; Lee, Y.; Kwak, J.; Lee, S.S. Increase in detection sensitivity of surface acoustic wave biosensor using triple transit echo wave. Appl. Phys. Lett. 2018, 113, 083702. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Yatsuda, H.; Goto, M.; Kondoh, J.; Liu, S.-H.; Wang, R.Y.L. Application of Shear Horizontal Surface Acoustic Wave (SH-SAW) Immunosensor in Point-of-Care Diagnosis. Biosensors 2023, 13, 605. [Google Scholar] [CrossRef] [PubMed]
- Maslova, A.O.; Hsing, I.M. Thiol-free oligonucleotide surface modification of gold nanoparticles for nanostructure assembly. Nanoscale Adv. 2019, 1, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Branch, D.W.; Brozik, S.M. Low-level detection of a Bacillus anthracis simulant using Love-wave biosensors on 36°YX LiTaO3. Curr. Opin. Biotechnol. 2023, 79, 102879. [Google Scholar]
- Schwesig, D.; Borchers, U.; Chancerelle, L.; Duffek, A.; Eriksson, U.; Goksøyr, A.; Lamoree, M.; Lepom, P.P.; Leonards, P.; Leverett, D.; et al. Network of reference la-boratories and related organisations for monitoring and bio monitoring of emerging environmental pollutants, Protocol for the Validation of Chemical and Biological Biomonitoring Methods. 2009. Available online: https://www.norman-network.net/sites/default/files/norman_v1_v2_v3_version_02_final_feb2009.pdf (accessed on 7 August 2024).
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef] [PubMed]
- Kumari, K.; Kar, A.; Nayak, A.K.; Mishra, S.K.; Subudhi, U. miRNA-mediated alteration of sulfatase modifying factor 1 expression using self-assembled branched DNA nanostructures. RSC Adv. 2021, 11, 10670–10680. [Google Scholar] [CrossRef]
- Yang, Y.; Chaerkady, R.; Beer, M.A.; Mendell, J.T.; Pandey, A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 2009, 9, 1374–1384. [Google Scholar] [CrossRef] [PubMed]



| Oligonucleotides | Sequences |
|---|---|
| miR-21 (control) | 5′-UAG CUU AUC AGA CUG AUG UUG A-3′ |
| miR-106b (target) | 5′-UAA AGU GCU GAC AGU GCA GAU-3′ |
| miR-124 (control) | 5′-CGU GUU CAC AGC GGA CCU UGA U-3′ |
| miR-155 (control) | 5′-UUA AUG CUA AUC GUG AUA GGG GUU-3′ |
| capture probes (complementary sequence of miR-106b) | 5′-H2N-(CH2)6-ATC TGC ACT GTC AGC ACT TTA CCC GGG CCC G-3′ |
| reference capture probe | 5′-H2N-(CH2)6-CCC CCC CCC C-3′ |
| target detecting probe | 5′-AAA AAA AAA A-(CH2)6-S-S-(CH2)6-OH-3′ |
| reference detecting probe | 5′-GGG GGG GGG G-(CH2)6-S-S-(CH2)6-OH-3′ |
| Methods | Target | Linear Range (pM) | LOD (pM) | References |
|---|---|---|---|---|
| Fluorescence | miR-21 | 0.0005–100 | 0.00017 | [19] |
| miR-21 | 5–5000 | 1.4 | [20] | |
| miR-155 | 10–400 | 5.0 | [21] | |
| Electrochemical | miR-21 | 0.001–1000 | 0.00002 | [22] |
| miR-34a | 100–1,000,000 | 0.00094 | [23] | |
| miR-224 | 1.0–1000 | 50 | [24] | |
| OECT | miR-21 | 0.001–100,000 | 0.01 | [25] |
| miR-21 | 5–20,000 | 2.0 | [26] | |
| QCM | miR-21 | 1000–10,000 | 400 | [27] |
| SPR | miR-155 | 1.0–1,000,000 | 0.5 | [28] |
| SERS | miR-200a | 0.0001–1000 | 0.00026 | [29] |
| SAW | miR-106b | 0.1–1,000,000 | 0.0034 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.B.; Lee, S.S. Isolation and Characterization of Exosomes from Cancer Cells Using Antibody-Functionalized Paddle Screw-Type Devices and Detection of Exosomal miRNA Using Piezoelectric Biosensor. Sensors 2024, 24, 5399. https://doi.org/10.3390/s24165399
Han SB, Lee SS. Isolation and Characterization of Exosomes from Cancer Cells Using Antibody-Functionalized Paddle Screw-Type Devices and Detection of Exosomal miRNA Using Piezoelectric Biosensor. Sensors. 2024; 24(16):5399. https://doi.org/10.3390/s24165399
Chicago/Turabian StyleHan, Su Bin, and Soo Suk Lee. 2024. "Isolation and Characterization of Exosomes from Cancer Cells Using Antibody-Functionalized Paddle Screw-Type Devices and Detection of Exosomal miRNA Using Piezoelectric Biosensor" Sensors 24, no. 16: 5399. https://doi.org/10.3390/s24165399
APA StyleHan, S. B., & Lee, S. S. (2024). Isolation and Characterization of Exosomes from Cancer Cells Using Antibody-Functionalized Paddle Screw-Type Devices and Detection of Exosomal miRNA Using Piezoelectric Biosensor. Sensors, 24(16), 5399. https://doi.org/10.3390/s24165399






