Evaluating MIR and NIR Spectroscopy Coupled with Multivariate Analysis for Detection and Quantification of Additives in Tobacco Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tobacco Samples
2.2. Reagents and Chemicals
2.3. Sample Preparation
2.4. Data Acquisition
2.4.1. MIR
2.4.2. NIR
2.4.3. Data Preprocessing
2.4.4. Selection of Training and Test Sets
2.4.5. Principal Component Analysis
2.4.6. Hierarchical Cluster Analysis
2.4.7. Soft Independent Modeling of Class Analogy
2.4.8. Partial Least Squares
2.5. Software for Data Analysis
3. Results and Discussion
3.1. Unsupervised Classification Models
3.1.1. PCA
3.1.2. HCA
3.2. Supervised Classification Models
3.2.1. SIMCA
3.2.2. PLS-DA Model
3.3. Quantitative Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanchez-Ramos, J.R. The Rise and Fall of Tobacco as a Botanical Medicine. J. Herb. Med. 2020, 22, 100374. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.W.; Cummings, K.M. Tobacco and Lung Cancer: Risks, Trends, and Outcomes in Patients with Cancer. Am. Soc. Clin. Oncol. Educ. Book 2013, 33, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Lickint, F. Tabak und Tabakrauch als Ätiologischer Faktor des Carcinoms. Z. Für Krebsforsch. 1930, 30, 349–365. [Google Scholar] [CrossRef]
- Marquardt, H.; Schäfer, S.G.; Barth, H. Toxikologie, 4th ed.; Wissenschaftliche Verlagsgesellschaft Stuttgart: Stuttgart, Germany, 2019; pp. 911–936. [Google Scholar]
- Rodgman, A.; Perfetti, T.A. The Chemical Components of Tobacco and Tobacco Smoke; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Rodgeman, A. Some Studies of the Effect of Additives on Cigarette Mainstream Smoke Properties. II. Casing Materials and Humectants. Contrib. Tob. Nicotine Res. 2002, 20, 279–299. [Google Scholar] [CrossRef]
- Roemer, E.; Schorp, M.K.; Piadé, J.-J.; Seeman, J.I.; Leyden, D.E.; Haussmann, H.-J. Scientific Assessment of the Use of Sugars as Cigarette Tobacco Ingredients: A Review of Published and Other Publicly Available Studies. Crit. Rev. Toxicol. 2012, 42, 244–278. [Google Scholar] [CrossRef]
- Simms, L.; Clarke, A.; Paschke, T.; Manson, A.; Murphy, J.; Stabbert, R.; Esposito, M.; Ghosh, D.; Roemer, E.; Martinez, J.; et al. Assessment of Priority Tobacco Additives per the Requirements of the EU Tobacco Products Directive (2014/40/EU): Part 1: Background, Approach, and Summary of Findings. Regul. Toxicol. Pharmacol. 2019, 104, 84–97. [Google Scholar] [CrossRef]
- Paumgartten, F.J.R.; Gomes-Carneiro, M.R.; de Oliveira, A.C.A.X. The Impact of Tobacco Additives on Cigarette Smoke Toxicity: A Critical Appraisal of Tobacco Industry Studies. Cad. Saúde Pública 2017, 33 (Suppl. S3), e00132415. [Google Scholar] [CrossRef]
- World Health Organization. WHO Framework Convention on Tobacco Control: Guidelines for Implementation Article 5.3; Article 8; Articles 9 and 10 Article 11; Article 12; Article 13; Article 14; WHO Library Cataloguing-in-Publication Data; World Health Organization: Geneva, Switzerland, 2013; Available online: http://apps.who.int/iris/bitstream/10665/80510/1/9789241505185_eng.pdf?ua=1 (accessed on 28 August 2022).
- European Commission. Directive 2014/40/EU of the European parliament and of the Council of 3 April 2014 on the approximation of the laws, regulations and administrative provisions of the Member States concerning the manufacture, presentation and sale of tobacco and related products and repealing Directive 2001/37/EC. Off. J. Eur. Union 2014, 127, 1–38. [Google Scholar]
- Nguyen, H.; Dennehy, C.E.; Tsourounis, C. Violation of US Regulations Regarding Online Marketing and Sale of E-Cigarettes: FDA Warnings and Retailer Responses. Tob. Control 2020, 29, e4–e9. [Google Scholar] [CrossRef]
- Jawad, M.; El Kadi, L.; Mugharbil, S.; Nakkash, R. Waterpipe Tobacco Smoking Legislation and Policy Enactment: A Global Analysis. Tob. Control 2015, 24 (Suppl. S1), i60–i65. [Google Scholar] [CrossRef]
- Hemmerich, N.; Jenson, D.; Bowrey, B.L.; Lee, J.G.L. Underutilisation of Notobacco-Sale Orders Against Retailers that Repeatedly Sell to Minors, 2015–2019, USA. Tob. Control 2022, 31, e99–e103. [Google Scholar] [CrossRef] [PubMed]
- Levinson, A.H.; Patnaik, J.L. A Practical Way to Estimate Retail Tobacco Sales Violation Rates More Accurately. Nicotine Tob. Res. 2013, 15, 1952–1955. [Google Scholar] [CrossRef] [PubMed]
- Marques, P.; Piqueras, L.; Sanz, M.J. An Updated Overview of E-Cigarette Impact on Human Health. Respir. Res. 2021, 22, 151. [Google Scholar] [CrossRef] [PubMed]
- Rehan, H.S.; Maini, J.; Hungin, A.P.S. Vaping Versus Smoking: A Quest for Efficacy and Safety of E-Cigarettes. Curr. Drug Saf. 2018, 13, 92–101. [Google Scholar] [CrossRef]
- Feeney, S.; Rossetti, V.; Terrien, J. E-Cigarettes: A Review of the Evidence—Harm Versus Harm Reduction. Tob. Use Insights 2022, 15, 1179173x221087524. [Google Scholar] [CrossRef]
- Travis, N.; Knoll, M.; Cadham, C.J.; Cook, S.; Warner, K.E.; Fleischer, N.L.; Douglas, C.E.; Sánchez-Romero, L.M.; Mistry, R.; Meza, R.; et al. Health Effects of Electronic Cigarettes: An Umbrella Review and Methodological Considerations. Int. J. Environ. Res. Public Health 2022, 19, 9054. [Google Scholar] [CrossRef]
- Marco, E.; Grimalt, J.O. A Rapid Method for the Chromatographic Analysis of Volatile Organic Compounds in Exhaled Breath of Tobacco Cigarette and Electronic Cigarette Smokers. J. Chromatogr. A 2015, 1410, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Nie, B.; Liu, T.; Yuan, F.; Feng, F.; Zhang, Y.; Zhou, W.; Xu, X.; Yao, M.; Zhang, F. Simultaneous Determination of Coumarin and Its Derivatives in Tobacco Products by Liquid Chromatography-Tandem Mass Spectrometry. Molecules 2016, 21, 1511. [Google Scholar] [CrossRef]
- Fekhar, M.; Daghbouche, Y.; Bouzidi, N.; El Hattab, M. ATR-MIR Spectroscopy Combined with Chemometrics for Quantification of Total Nicotine in Algerian Smokeless Tobacco Products. Microchem. J. 2023, 193, 109127. [Google Scholar] [CrossRef]
- Duell, A.K.; Pankow, J.F.; Peyton, D.H. Free-Base Nicotine Determination in Electronic Cigarette Liquids by 1H NMR Spectroscopy. Chem. Res. Toxicol. 2018, 31, 431–434. [Google Scholar] [CrossRef]
- Kapar, A.; Ibraimov, A.B.; Sergazina, M.M.; Alimzhanova, M.B.; Abilev, M.B. Analysis of Tobacco Products by Chromatography Methods. Int. J. Biol. Chem. 2018, 11, 133–141. [Google Scholar]
- Brima, E.I. Determination of Metal Levels in Shamma (Smokeless Tobacco) with Inductively Coupled Plasma Mass Spectrometry (ICP-MS) in Najran, Saudi Arabia. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 4761. [Google Scholar]
- Cheng, D.; Ni, Z.; Liu, M.; Shen, X.; Jia, Y. Determination of Trace Cr, Ni, Hg, As, and Pb in the Tipping Paper and Filters of Cigarettes by Monochromatic Wavelength X-Ray Fluorescence Spectrometry. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2021, 502, 59–65. [Google Scholar] [CrossRef]
- Gómez-Siurana, A.; Marcilla, A.; Beltran, M.; Martinez, I.; Berenguer, D.; García-Martínez, R.; Hernández-Selva, T. Thermogravimetric Study of the Pyrolysis of Tobacco and Several Ingredients Used in the Fabrication of Commercial Cigarettes: Effect of the Presence of MCM-41. Thermochim. Acta 2011, 523, 161–169. [Google Scholar] [CrossRef]
- Merckel, C.; Pragst, F.; Ratzinger, A.; Aebi, B.; Bernhard, W.; Sporkert, F. Application of Headspace Solid Phase Microextraction to Qualitative and Quantitative Analysis of Tobacco Additives in Cigarettes. J. Chromatogr. A 2006, 1116, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, K.; David, F.; Dumont, E.; Ochiai, N.; Tamura, H.; Sandra, P. LC Fractionation Followed by Pyrolysis GC–MS for the In-Depth Study of Aroma Compounds Formed During Tobacco Combustion. J. Anal. Appl. Pyrolysis 2015, 116, 68–74. [Google Scholar] [CrossRef]
- Caruso, R.V.; O’Connor, R.J.; Stephens, W.E.; Fong, G.T. Tobacco Industry Manipulation of Menthol Content in Cigarettes and Population Menthol Preferences: A Review of Tobacco Industry Documents. Tob. Control 2019, 28, 295–302. [Google Scholar]
- Heckman, C.A.; Meeker, R.J. Cocoa and Chocolate in Human Health and Disease. Antioxid. Redox Signal. 2011, 15, 2779–2811. [Google Scholar]
- Stanfill, S.B.; Jia, L.T.; Ashley, D.L.; Watson, C.H. Surveillance of Caffeine in Cigarette Tobacco. Food Chem. Toxicol. 2003, 41, 1297–1302. [Google Scholar]
- European Directorate for the Quality of Medicines. Qualification of Equipment, Annex 4: Qualification of IR Spectrophotometers (PA/PH/OMCL (07) 12 DEF CORR). 2007. [Google Scholar]
- McClure, W.F. Near-Infrared Spectroscopy: The Giant Is Running Strong. Anal. Chem. 1994, 66, 42A–53A. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Snee, R.D. Validation of Regression Models: Methods and Examples. Technometrics 1977, 19, 415–428. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Abdi, H.; Williams, L.J. Principal Component Analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Altman, N.S. An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. Am. Stat. 1992, 46, 175–185. [Google Scholar] [CrossRef]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS Discriminant Analysis: Combining the Strengths of PLS-DA and SIMCA Classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Candolfi, A.; Massart, D.L.; Heuerding, S. Decision Criteria for Soft Independent Modelling of Class Analogy Applied to near Infrared Data. Chemom. Intell. Lab. Syst. 1999, 47, 65–77. [Google Scholar] [CrossRef]
- Gurbanov, R.; Gozen, A.G.; Severcan, F. Rapid Classification of Heavy Metal-Exposed Freshwater Bacteria by Infrared Spectroscopy Coupled with Chemometrics Using Supervised Method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 282–290. [Google Scholar] [CrossRef]
- Saleh, A.A.; Hegazy, M.; Abbas, S.; Elkosasy, A. Development of Distribution Maps of Spectrally Similar Degradation Products by Raman Chemical Imaging Microscope Coupled with a New Variable Selection Technique and SIMCA Classifier. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 268, 120654. [Google Scholar] [CrossRef]
- Kemsley, E.K. Discriminant Analysis of High-Dimensional Data: A Comparison of Principal Components Analysis and Partial Least Squares Data Reduction Methods. Chemom. Intell. Lab. Syst. 1996, 33, 47–61. [Google Scholar] [CrossRef]
- Chevallier, S.; Bertrand, D.; Kohler, A.; Courcoux, P. Application of PLS-DA in Multivariate Image Analysis. J. Chemom. 2006, 20, 221–229. [Google Scholar] [CrossRef]
- Akhtar, Z.; Barhdadi, S.; De Braekeleer, K.; Delporte, C.; Adams, E.; Deconinck, E. Spectroscopy and Chemometrics for Conformity Analysis of E-Liquids: Illegal Additive Detection and Nicotine Characterization. Chemosensors 2024, 12, 9. [Google Scholar] [CrossRef]
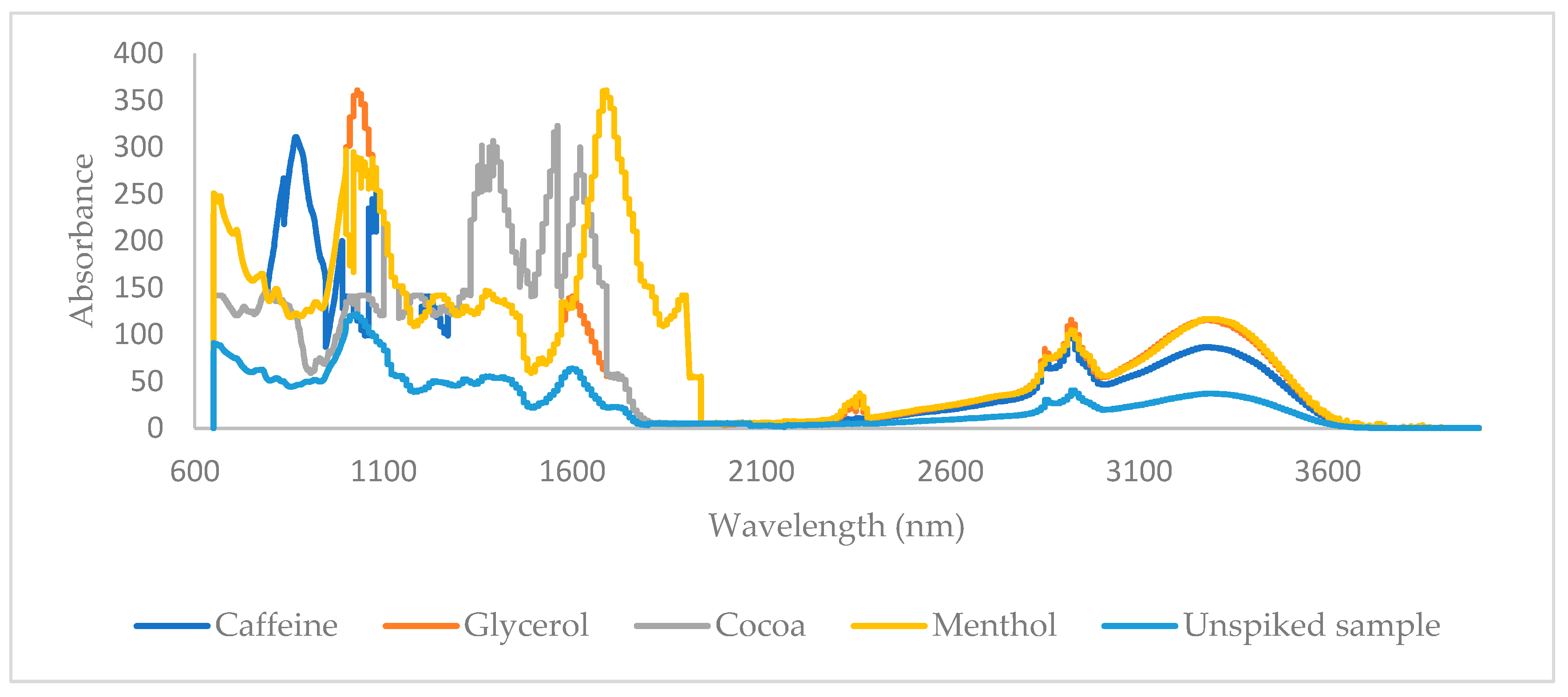
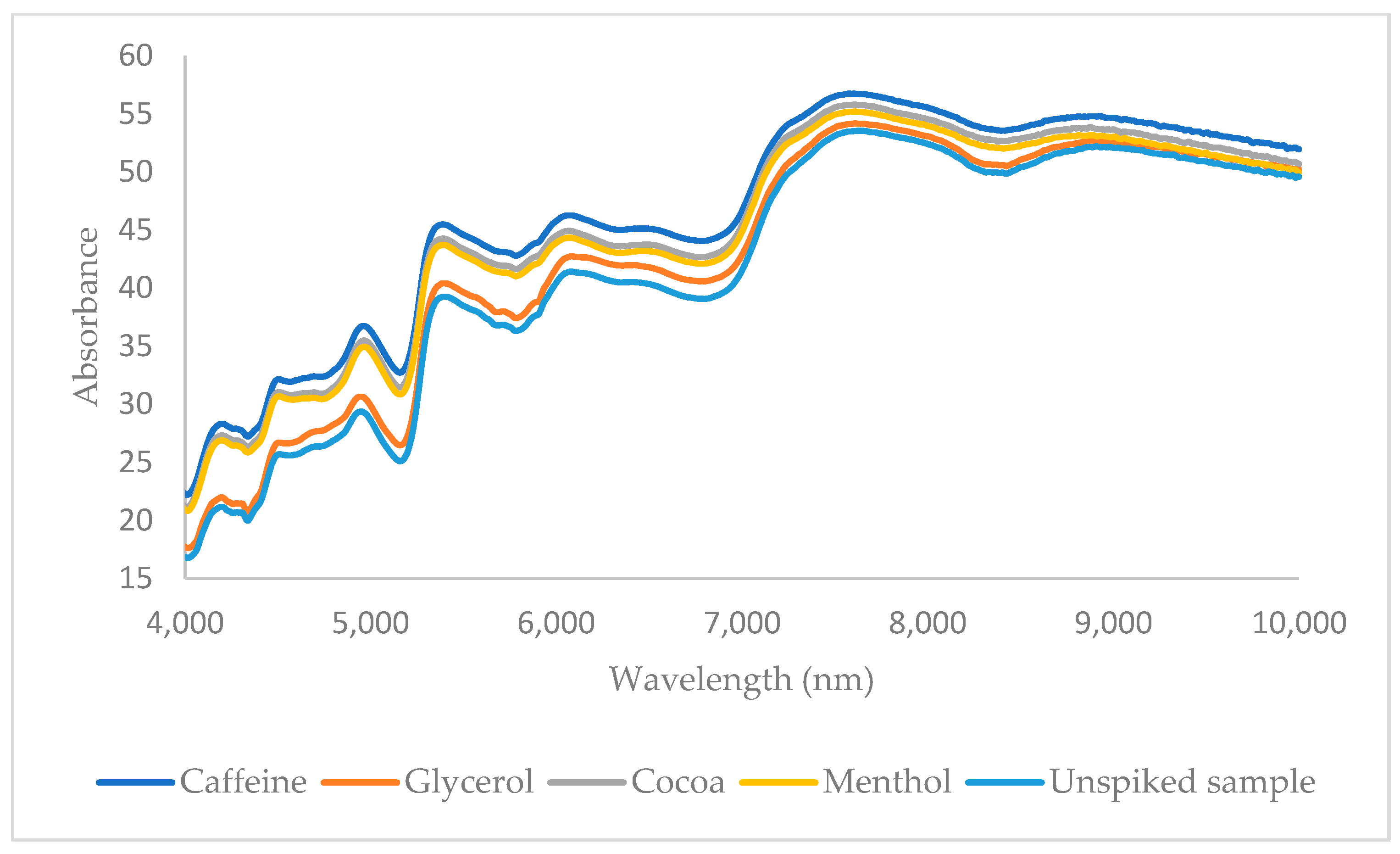





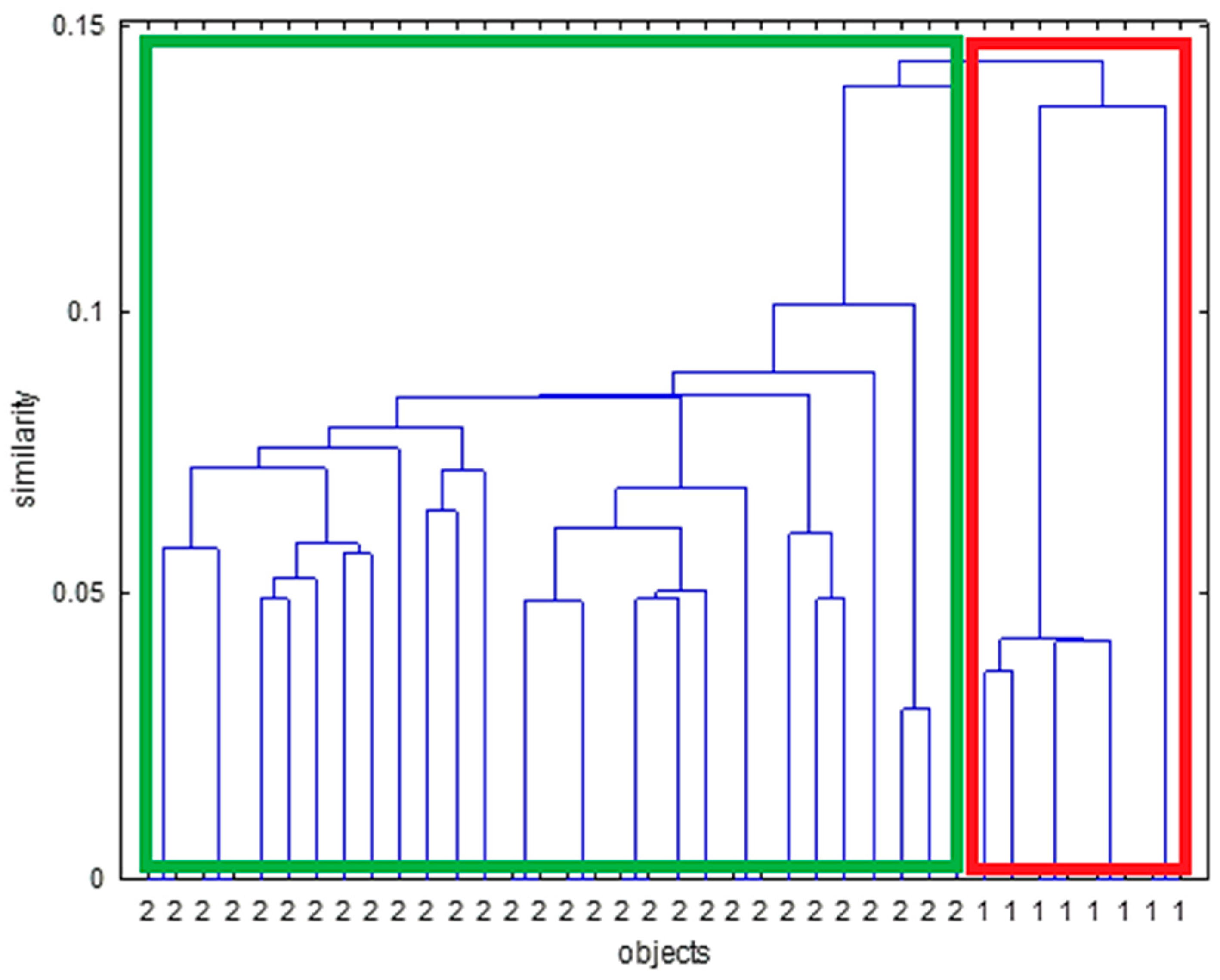
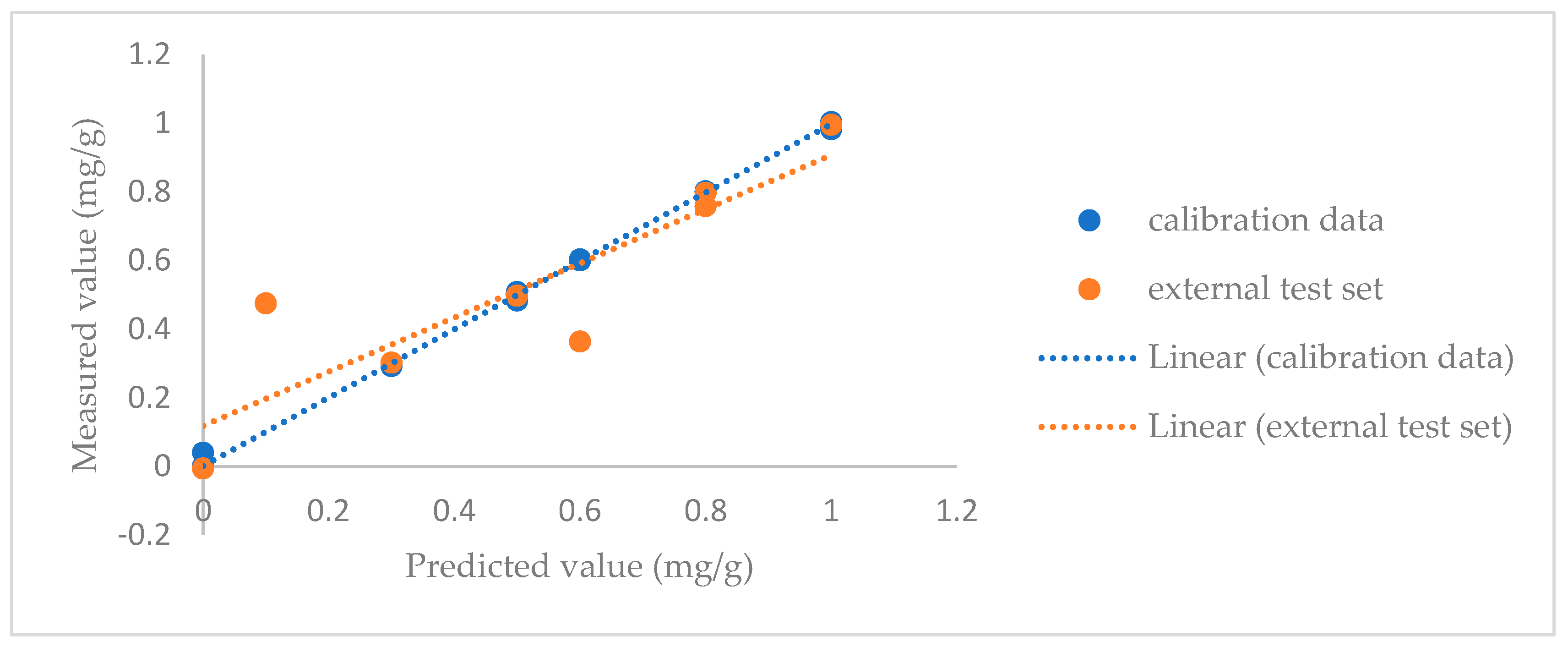
| Spectroscopic Technique | Data Pretreatment | Chemometric Technique | Target Molecule | No. of PCs | No. of Samples in External Test Set | Correct Classification Rate (External Test Set) [No. of Negative Samples] | No. of Samples in the Training Set | Correct Classification Rate (Cross-Validation) [No. of Negative Samples] |
|---|---|---|---|---|---|---|---|---|
| MIR | SNV | Caffeine | 3-2 | 8 | 88% (7/8) [2/8] | 30 | 97% (29/30) [5/30] | |
| Autoscaling | Glycerol | 3-1 | 8 | 100% (8/8) [2/8] | 30 | 90% (27/30) [5/30] | ||
| 1st derivative | SIMCA | Menthol | 2-1 | 8 | 88% (7/8) [3/8] | 30 | 97% (29/30) [4/30] | |
| Autoscaling | Cocoa | 3-2 | 8 | 88% (7/8) [2/8] | 30 | 90% (27/30) [5/30] | ||
| NIR | SNV | Caffeine | 2-1 | 8 | 100% (8/8) [3/8] | 30 | 100% (30/30) [4/30] | |
| Autoscaling | Glycerol | 1-1 | 8 | 88% (7/8) [2/8] | 30 | 97% (29/30) [5/30] | ||
| Autoscaling | SIMCA | Menthol | 2-1 | 8 | 100% (8/8) [2/8] | 30 | 100% (30/30) [5/30] | |
| Autoscaling | Cocoa | 2-2 | 8 | 100% (8/8) [3/8] | 30 | 97% (29/30) [4/30] |
| SIMCA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIR | Caffeine | Glycerol | Menthol | Cocoa | ||||||||
| Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | |
| Cross-validation | 0.96 | 0.85 | 1.00 | 0.92 | 0.75 | 0.66 | 0.96 | 0.85 | 1.00 | 0.96 | 0.85 | 0.66 |
| Test set | 0.87 | 0.50 | 1.00 | 1.00 | 1.00 | 1.00 | 0.87 | 0.50 | 1.00 | 0.87 | 0.50 | 1.00 |
| NIR | Caffeine | Glycerol | Menthol | Cocoa | ||||||||
| Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | |
| Cross-validation | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 1.00 | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 1.00 |
| Test set | 1.00 | 1.00 | 1.00 | 0.87 | 0.50 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Spectroscopic Technique | Data Pretreatment | Chemometric Technique | Target Molecule | No of Latent Variables | No of Samples in External Test Set | Correct Classification Rate (External Test Set) [No. of Negative Samples] | No of Samples in Training Set | Correct Classification Rate (Cross-Validation) [No. of Negative Samples] |
|---|---|---|---|---|---|---|---|---|
| MIR | 1st derivative | Caffeine | 4 | 8 | 100% (8/8) [3/8] | 30 | 100% (30/30) [4/30] | |
| Autoscaling | Glycerol | 2 | 8 | 100% (8/8) [2/8] | 30 | 93% (28/30) [5/30] | ||
| 2nd derivative | PLS-DA | Menthol | 7 | 8 | 100% (8/8) [3/8] | 30 | 93% (28/30) [4/30] | |
| 1st derivative | Cocoa | 4 | 8 | 88% (7/8) [2/8] | 30 | 90% (27/30) [5/30] | ||
| NIR | Autoscaling | Caffeine | 2 | 8 | 100% (8/8) [2/8] | 30 | 100% (30/30) [5/30] | |
| Autoscaling | Glycerol | 6 | 8 | 100% (8/8) [2/8] | 30 | 93% (28/30) [5/30] | ||
| Autoscaling | PLS-DA | Menthol | 4 | 8 | 88% (7/8) [2/8] | 30 | 97% (29/30) [5/30] | |
| 1st derivative | Cocoa | 8 | 8 | 100% (8/8) [3/8] | 30 | 80% (24/30) [4/30] |
| PLS-DA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIR | Caffeine | Glycerol | Menthol | Cocoa | ||||||||
| Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | |
| Cross-validation | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 0.50 | 0.92 | 0.75 | 1.00 | 0.96 | 0.85 | 0.66 |
| Test set | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 1.00 |
| NIR | Caffeine | Glycerol | Menthol | Cocoa | ||||||||
| Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | Precision | Specificity | Sensitivity | |
| Cross-validation | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 0.50 | 0.96 | 0.85 | 1.00 | 0.85 | 0.60 | 0.66 |
| Test set | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.96 | 0.85 | 1.00 | 1.00 | 1.00 | 1.00 |
| Spectroscopic Technique | Data Pretreatment | Target Molecule | No. of Latent Variables | RMSEC | R2c | RMSEP | R2p | RMSECV | R²cv |
|---|---|---|---|---|---|---|---|---|---|
| MIR | SNV | Caffeine | 9 | 0.0088 | 0.9983 | 0.1514 | 0.7781 | 0.1634 | 0.7802 |
| 2nd derivative | Glycerol | 12 | 0.1541 | 0.7331 | 0.1931 | 0.6995 | 0.1923 | 0.7025 | |
| 1st derivative | Menthol | 8 | 0.0670 | 0.9582 | 0.1601 | 0.7710 | 0.1580 | 0.7993 | |
| 1st derivative | Cocoa | 8 | 0.1873 | 0.8019 | 0.2074 | 0.7195 | 0.1750 | 0.7584 | |
| NIR | 1st derivative | Caffeine | 10 | 0.0139 | 0.9983 | 0.1453 | 0.7138 | 0.1079 | 0.9141 |
| 1st derivative | Glycerol | 9 | 0.1078 | 0.8765 | 0.2052 | 0.6926 | 0.1700 | 0.7667 | |
| Autoscaling | Menthol | 13 | 0.0806 | 0.9443 | 0.3119 | 0.7572 | 0.1675 | 0.7738 | |
| SNV | Cocoa | 12 | 0.3112 | 0.7993 | 0.2816 | 0.6435 | 0.1900 | 0.7190 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhtar, Z.; Canfyn, M.; Vanhee, C.; Delporte, C.; Adams, E.; Deconinck, E. Evaluating MIR and NIR Spectroscopy Coupled with Multivariate Analysis for Detection and Quantification of Additives in Tobacco Products. Sensors 2024, 24, 7018. https://doi.org/10.3390/s24217018
Akhtar Z, Canfyn M, Vanhee C, Delporte C, Adams E, Deconinck E. Evaluating MIR and NIR Spectroscopy Coupled with Multivariate Analysis for Detection and Quantification of Additives in Tobacco Products. Sensors. 2024; 24(21):7018. https://doi.org/10.3390/s24217018
Chicago/Turabian StyleAkhtar, Zeb, Michaël Canfyn, Céline Vanhee, Cédric Delporte, Erwin Adams, and Eric Deconinck. 2024. "Evaluating MIR and NIR Spectroscopy Coupled with Multivariate Analysis for Detection and Quantification of Additives in Tobacco Products" Sensors 24, no. 21: 7018. https://doi.org/10.3390/s24217018
APA StyleAkhtar, Z., Canfyn, M., Vanhee, C., Delporte, C., Adams, E., & Deconinck, E. (2024). Evaluating MIR and NIR Spectroscopy Coupled with Multivariate Analysis for Detection and Quantification of Additives in Tobacco Products. Sensors, 24(21), 7018. https://doi.org/10.3390/s24217018






