Detection of Biomolecules Using Solid-State Nanopores Fabricated by Controlled Dielectric Breakdown
Abstract
:1. Introduction
2. Materials and Methods
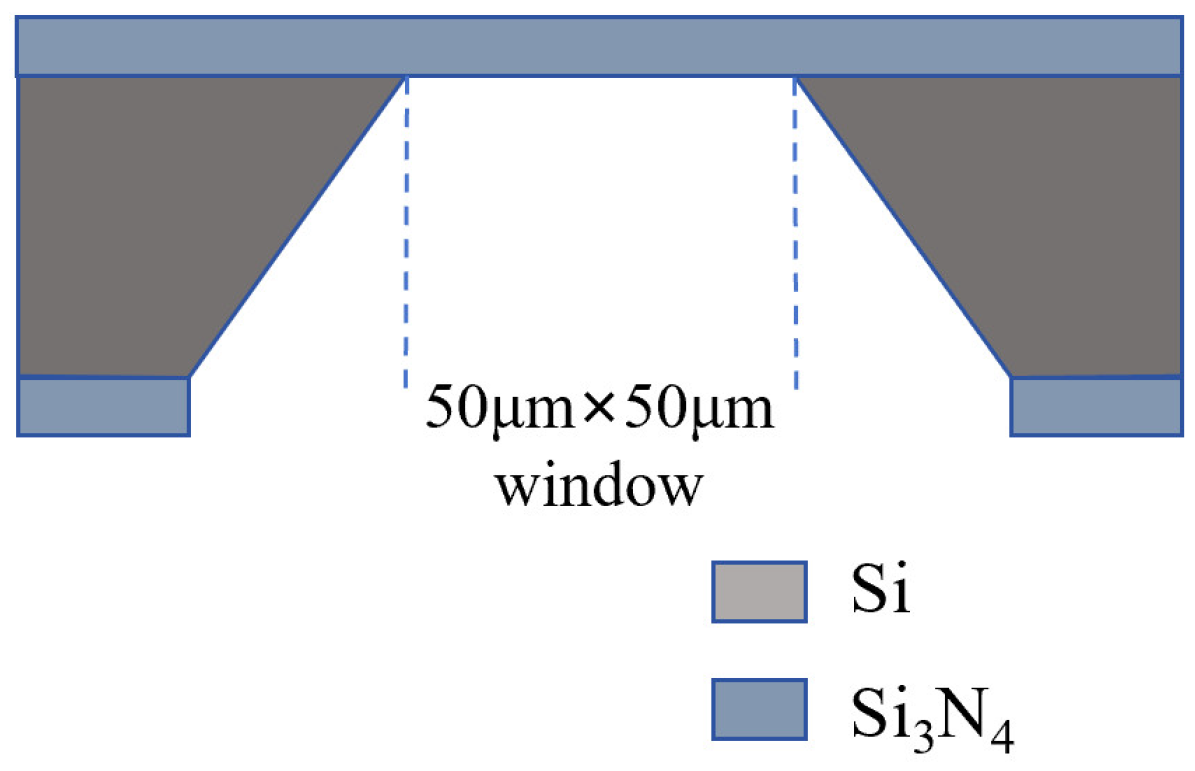
2.1. Nanopore Fabrication
2.2. Nanopore Characterization
2.3. Biomolecule Detection Experiment
3. Results
3.1. Characterization Results of Nanopores
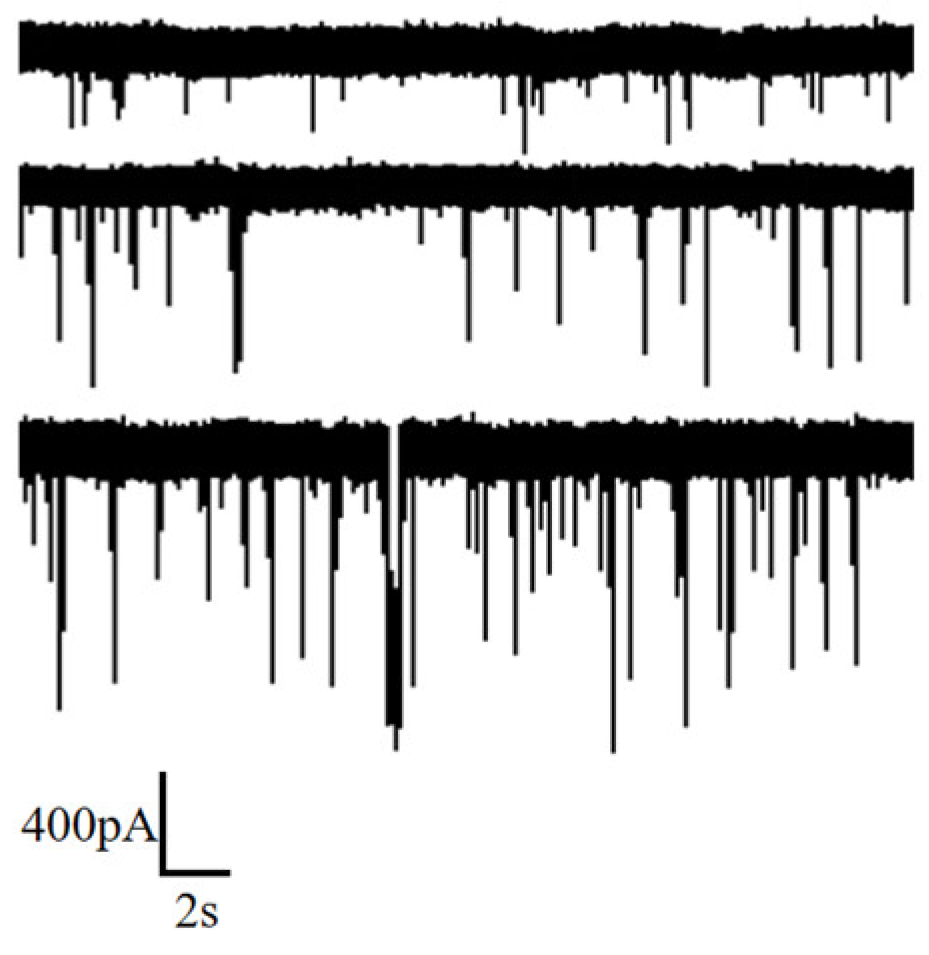
3.2. DNA Detection by Nanopore
3.2.1. Individual Event Analysis
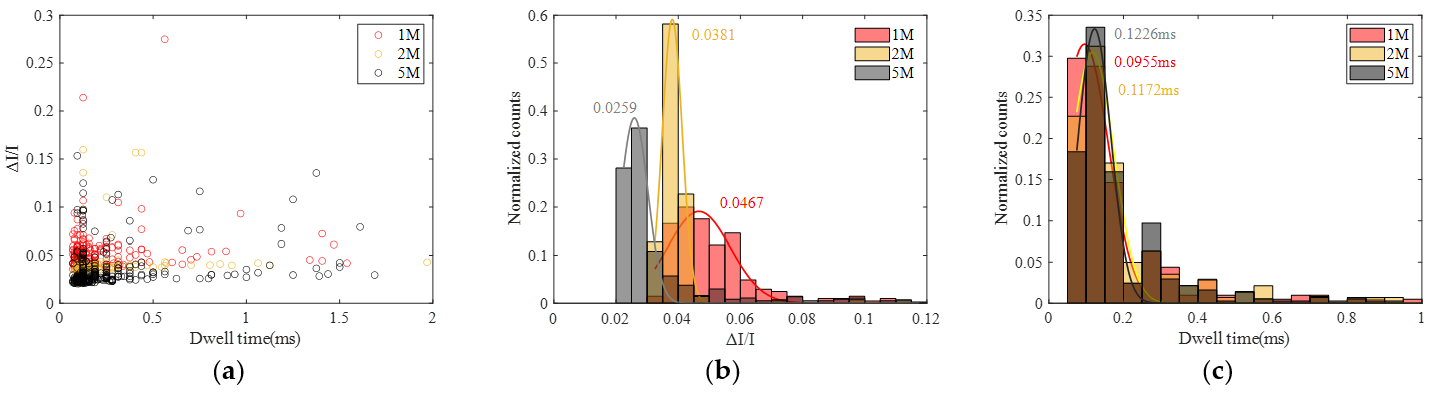
3.2.2. Experiment of Different Voltages
3.2.3. Experiment of Different Salt Concentration
3.2.4. Theoretical Analysis
3.3. Protein Detection by Nanopore
3.3.1. Experiment of Different Voltages
3.3.2. Experiment of Different Salt Concentration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, S. Nanopore-based sensing devices and applications to genome sequencing: A brief history and the missing pieces. Chin. Sci. Bull. 2014, 59, 4918–4928. [Google Scholar] [CrossRef]
- Marx, V. Nanopores: A sequencer in your backpack. Nat. Methods 2015, 12, 1015–1018. [Google Scholar] [CrossRef] [PubMed]
- Manrao, E.A.; Derrington, I.M.; Laszlo, A.H.; Langford, K.W.; Hopper, M.K.; Gillgren, N.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012, 30, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Bian, P.; Wang, H.; Sha, J. Detection of Different Structural Proteins Using Solid-state Nanopores. In Proceedings of the 2023 IEEE 18th International Conference on Nano/Micro Engineered and Molecular Systems (NEMS), Jeju Island, Republic of Korea, 14–17 May 2023. [Google Scholar]
- Xi, D.; Shang, J.; Fan, E.; You, J.; Zhang, S.; Wang, H. Nanopore-based selective discrimination of microRNAs with single-nucleotide difference using locked nucleic acid-modified probes. Anal. Chem. 2016, 88, 10540–10546. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Yamazaki, H.; Ren, R.; Wanunu, M.; Ivanov, A.P.; Edel, J.B. Solid-state nanopore sensors. Nat. Rev. Mater. 2020, 5, 931–951. [Google Scholar] [CrossRef]
- Wendell, D.; Jing, P.; Geng, J.; Subramaniam, V.; Lee, T.J.; Montemagno, C. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat. Nanotechnol. 2009, 4, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, C.; Zhang, Y.; Geng, J. Biological nanopores for sensing applications. Proteins 2022, 90, 1786–1799. [Google Scholar] [CrossRef]
- Bhatti, H.; Jawed, R.; Ali, I.; Iqbal, K.; Han, Y.; Lu, Z.; Liu, Q. Recent advances in biological nanopores for nanopore sequencing, sensing and comparison of functional variations in MspA mutants. RSC Adv. 2021, 11, 28996–29014. [Google Scholar] [CrossRef] [PubMed]
- Shekar, S.; Niedzwiecki, D.J.; Chien, C.-C.; Ong, P.; Fleischer, D.A.; Lin, J.; Rosenstein, J.K.; Drndić, M.; Shepard, K.L. Measurement of DNA translocation dynamics in a solid-state nanopore at 100 ns temporal resolution. Nano Lett. 2016, 16, 4483–4489. [Google Scholar] [CrossRef]
- Hu, R.; Rodrigues, J.V.; Waduge, P.; Yamazaki, H.; Cressiot, B.; Chishti, Y.; Makowski, L.; Yu, D.; Shakhnovich, E.; Zhao, Q.; et al. Differential enzyme flexibility probed using solid-state nanopores. ACS Nano 2018, 12, 4494–4502. [Google Scholar] [CrossRef]
- Carlsen, A.T.; Briggs, K.; Hall, A.R.; Tabard-Cossa, V. Solid-state nanopore localization by controlled breakdown of selectively thinned membranes. Nanotechnology 2017, 28, 085304. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-S.; Hong, S.C.; Wu, S.; Kim, H.-M.; Lee, C.; Lee, J.-S.; Lee, J.E.; Kim, K.-B. Differentiation of selectively labeled peptides using solid-state nanopores. Nanoscale 2019, 11, 2510–2520. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.W.; Lee, J.Y.; Oh, D.H.; Kim, T.W.; Cho, W.J. Shrinkage and expansion mechanisms of SiO2 elliptical membrane nanopores. Appl. Phys. Lett. 2008, 93, 221903. [Google Scholar] [CrossRef]
- Larkin, J.; Henley, R.Y.; Jadhav, V.; Korlach, J.; Wanunu, M. Length-independent DNA packing into nanopore zero-mode waveguides for low-input DNA sequencing. Nat. Nanotechnol. 2017, 12, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Q.; Deng, T.; Liu, Z. Self-aligned nanopore formed on a SiO2 pyramidal membrane by a multipulse dielectric breakdown method. J. Phys. Chem. C 2018, 122, 11516–11523. [Google Scholar] [CrossRef]
- Merchant, C.A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fischbein, M.D.; Venta, K.; Luo, Z.; Johnson, A.T.C.; et al. DNA translocation through graphene nanopores. Nano Lett. 2010, 10, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Huang, Q.; Zhao, Y.; Zhou, D.; Ying, C.; Wang, D. Precise fabrication of a 5 nm graphene nanopore with a helium ion microscope for biomolecule detection. Nanotechnology 2016, 28, 045302. [Google Scholar] [CrossRef] [PubMed]
- Goyal, G.; Lee, Y.B.; Darvish, A.; Ahn, C.W.; Kim, M.J. Hydrophilic and size-controlled graphene nanopores for protein detection. Nanotechnology 2016, 27, 495301. [Google Scholar] [CrossRef]
- Liu, S.; Lu, B.; Zhao, Q.; Li, J.; Gao, T.; Chen, Y.; Zhang, Y.; Liu, Z.; Fan, Z.; Yang, F.; et al. Boron nitride nanopores: Highly sensitive DNA single-molecule detectors. Adv. Mater. 2013, 25, 4549–4554. [Google Scholar] [CrossRef]
- Park, K.-B.; Kim, H.-J.; Kim, H.-M.; Han, S.A.; Lee, K.H.; Kim, S.-W.; Kim, K.-B. Noise and sensitivity characteristics of solid-state nanopores with a boron nitride 2-D membrane on a pyrex substrate. Nanoscale 2016, 8, 5755–5763. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Liu, K.; Bulushev, R.D.; Khlybov, S.; Dumcenco, D.; Kis, A. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotechnol. 2015, 10, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tsutsui, M.; Zhou, Y.; Miao, X.-S. Solid-state nanopore systems: From materials to applications. NPG Asia Mater. 2021, 13, 48. [Google Scholar] [CrossRef]
- Gierak, J.; Madouri, A.; Biance, A.L.; Bourhis, E.; Patriarche, G.; Ulysse, C.; Lucot, D.; Lafosse, X.; Auvray, L.; Bruchhaus, L.; et al. Sub-5 nm FIB direct patterning of nanodevices. Microelectron. Eng. 2007, 84, 779–783. [Google Scholar] [CrossRef]
- Rigo, E.; Dong, Z.; Park, J.H.; Kennedy, E.; Hokmabadi, M.; Almonte-Garcia, L.; Ding, L.; Aluru, N. Measurements of the size and correlations between ions using an electrolytic point contact. Nat. Commun. 2019, 10, 2382. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.; Briggs, K.; Tabard-Cossa, V. Nanopore fabrication by controlled dielectric breakdown. PLoS ONE 2014, 9, e92880. [Google Scholar] [CrossRef]
- Arcadia, C.E.; Reyes, C.C.; Rosenstein, J.K. In situ nanopore fabrication and single-molecule sensing with microscale liquid contacts. ACS Nano 2017, 11, 4907–4915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Miyahara, Y.; Derriche, N.; Yang, W.; Yazda, K.; Capaldi, X.; Liu, Z.; Grutter, P.; Reisner, W. Nanopore formation via tip-controlled local breakdown using an atomic force microscope. Small Methods 2019, 3, 1900147. [Google Scholar] [CrossRef]
- Waugh, M.; Briggs, K.; Gunn, D.; Gibeault, M.; King, S.; Ingram, Q.; Jimenez, A.M.; Berryman, S.; Lomovtsev, D.; Andrzejewski, L.; et al. Solid-state nanopore fabrication by automated controlled breakdown. Nat. Protoc. 2020, 15, 122–143. [Google Scholar] [CrossRef]
- Gu, Z.; Ma, D.; Zhang, Z.; Zhang, Y.; Sha, J. Fabrication of Small-Scale Solid-State Nanopores by Dielectric Breakdown. In Proceedings of the 2020 IEEE 15th International Conference on Nano/Micro Engineered and Molecular System (NEMS), San Diego, CA, USA, 27–30 September 2020. [Google Scholar]
- Yanagi, I.; Akahori, R.; Hatano, T.; Takeda, K.-I. Fabricating nanopores with diameters of sub-1 nm to 3 nm using multilevel pulse-voltage injection. Sci. Rep. 2014, 4, 5000. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tsutsui, M.; Fan, C.; Taniguchi, M.; Kawai, T. Controlling DNA translocation through gate modulation of nanopore wall surface charges. ACS Nano 2011, 5, 5509–5518. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qiu, Y.H.; Yuan, Z.S.; Zhang, Y.; Sha, J.; Liu, L.; Sun, L.; Ni, Z.; Yi, H.; Li, D.; et al. Detection of Short Single-Strand DNA Homopolymers with Ultrathin Si3N4 Nanopores. Phys. Rev. E. 2015, 92, 022719. [Google Scholar] [CrossRef] [PubMed]
- Oukhaled, A.; Cressiot, B.; Bacri, L.; Pastoriza-Gallego, M.; Betton, J.M.; Bourhis, E.; Jede, R.; Gierak, J.; Auvray, L.; Pelta, J. Dynamics of completely unfolded and native proteins through solid-state nanopores as a function of electric driving force. ACS Nano 2011, 5, 3628–3638. [Google Scholar] [CrossRef] [PubMed]
- Maity, H.; Muttathukattil, A.N.; Reddy, G. Salt Effects on Protein Folding Thermodynamics. J. Phys. Chem. Lett. 2018, 9, 5063–5070. [Google Scholar] [CrossRef] [PubMed]
- Fujinami Tanimoto, I.M.; Zhang, J.; Cressiot, B.; Le Pioufle, B.; Bacri, L.; Pelta, J. Dynamics of DNA through solid-state nanopores fabricated by controlled dielectric breakdown. Chem. Asian J. 2022, 17, e202200888. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, P.; Zhao, C.; Pan, Q.; Xiong, Z.; Chen, Q.; Miao, X.; He, Y. Detection of Biomolecules Using Solid-State Nanopores Fabricated by Controlled Dielectric Breakdown. Sensors 2024, 24, 2420. https://doi.org/10.3390/s24082420
Cheng P, Zhao C, Pan Q, Xiong Z, Chen Q, Miao X, He Y. Detection of Biomolecules Using Solid-State Nanopores Fabricated by Controlled Dielectric Breakdown. Sensors. 2024; 24(8):2420. https://doi.org/10.3390/s24082420
Chicago/Turabian StyleCheng, Peng, Candong Zhao, Qinjie Pan, Zijian Xiong, Qi Chen, Xiangshui Miao, and Yuhui He. 2024. "Detection of Biomolecules Using Solid-State Nanopores Fabricated by Controlled Dielectric Breakdown" Sensors 24, no. 8: 2420. https://doi.org/10.3390/s24082420




