Content-Based Histopathological Image Retrieval
Abstract
1. Introduction
- A unified model for extracting image descriptor embeddings from histopathological images using multi-scale local–global fused features trained with Sub-center ArcFace loss [11].
- Two novel fusion operations, called Local Aggregator and Global Aggregator, employing a channel attention mechanism to enhance local and global feature fusion.
- A validation of the proposed model using the state-of-the-art CBHIR dataset Kimia Patch24C [12], demonstrating improved Recall@1 through experiments with the proposed embeddings.
2. Related Works
3. Proposed Method
3.1. Motivation
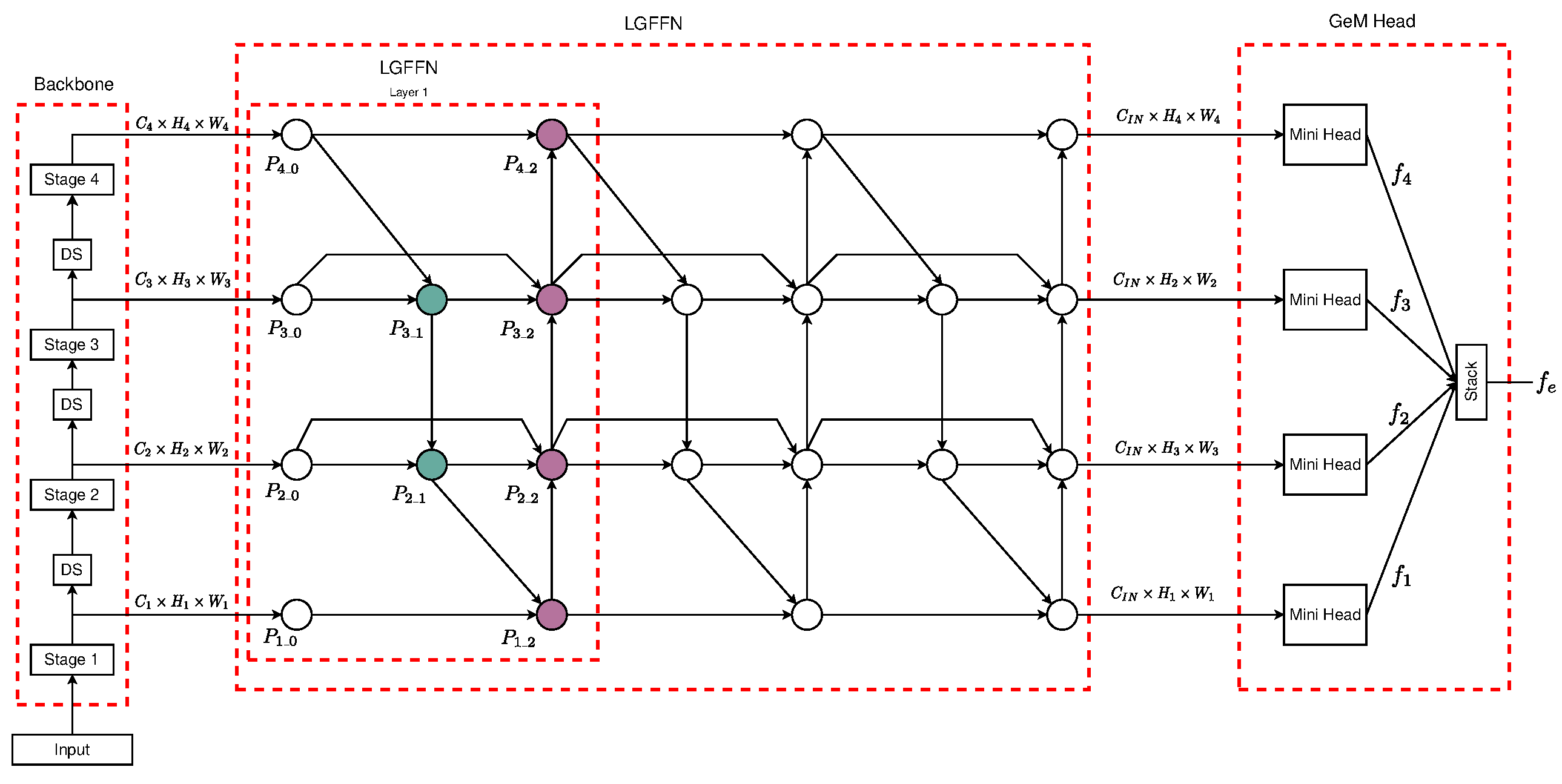
3.2. Local–Global Feature Fusion Embedding Model (LGFFEM)
3.3. Feature Fusion Neck
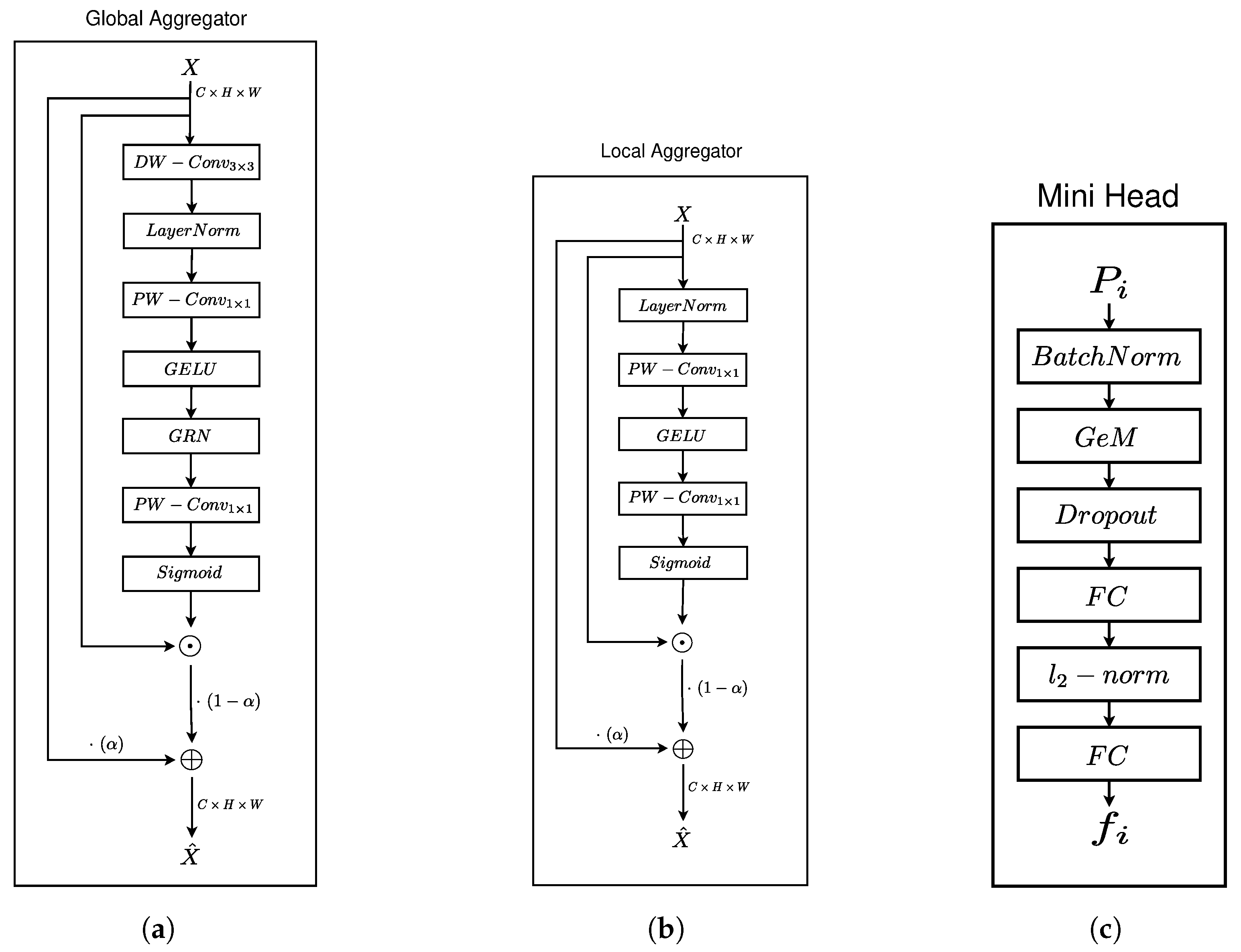
3.3.1. Feature Aggregator Units
- Local Feature: The given features have a minimum receptive field; these features preserve complex spatial information, thus facilitating the generation of high-level features.
- Global Feature: The features extracted from a generalization operation from an expansive receptive field are adept at capturing robust semantic information. These are categorized as low-level features.
Global Feature Aggregator
Local Feature Aggregator
3.3.2. Neck’s Architecture
3.4. Embedding Head
4. Experiments
4.1. Experimental Setup
4.1.1. Datasets and Evaluation Techniques
4.1.2. Backbone and Implementation Detail
4.1.3. Training Strategies
4.2. Results and Analysis
4.2.1. Metric Evaluation
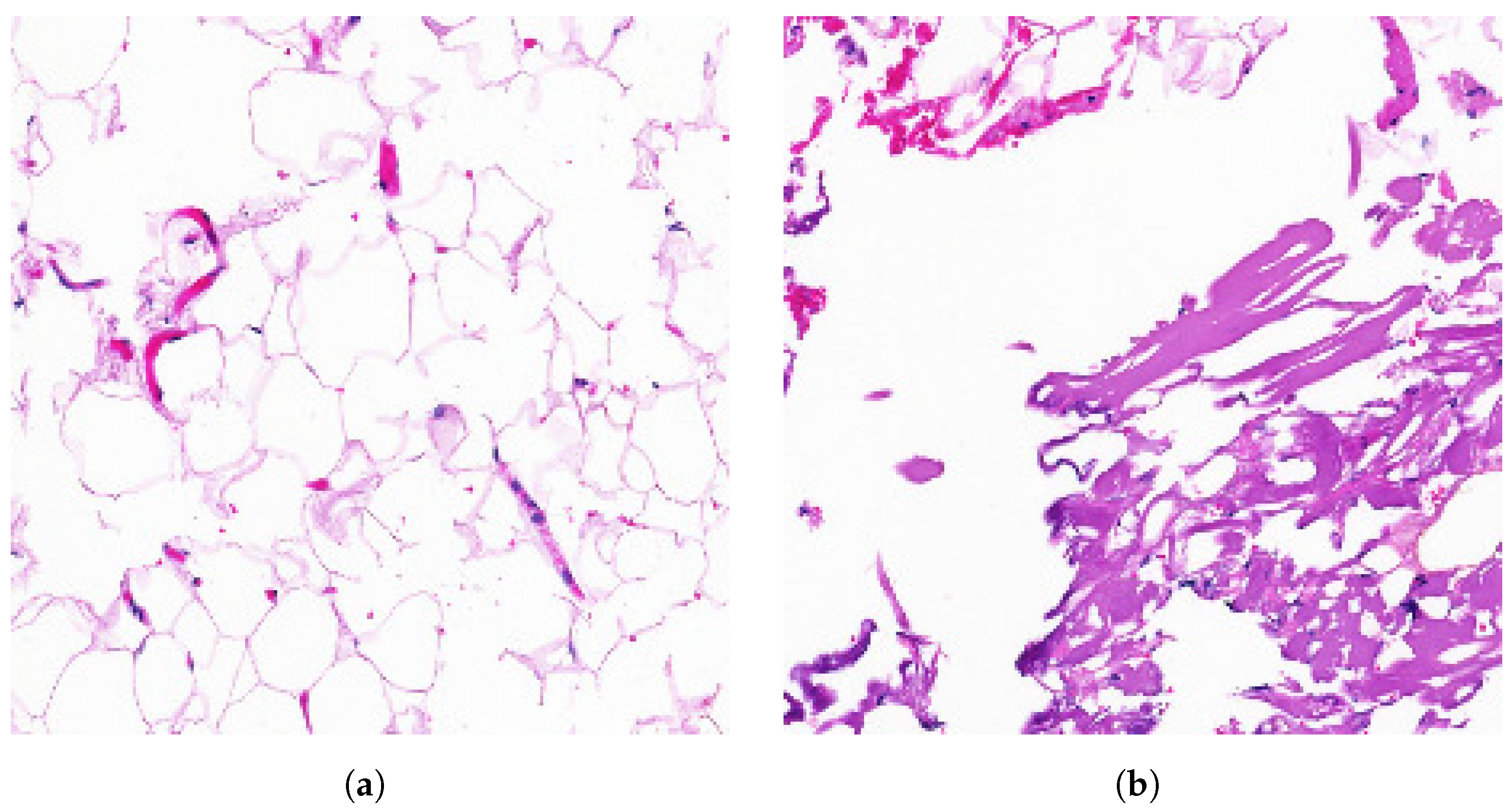
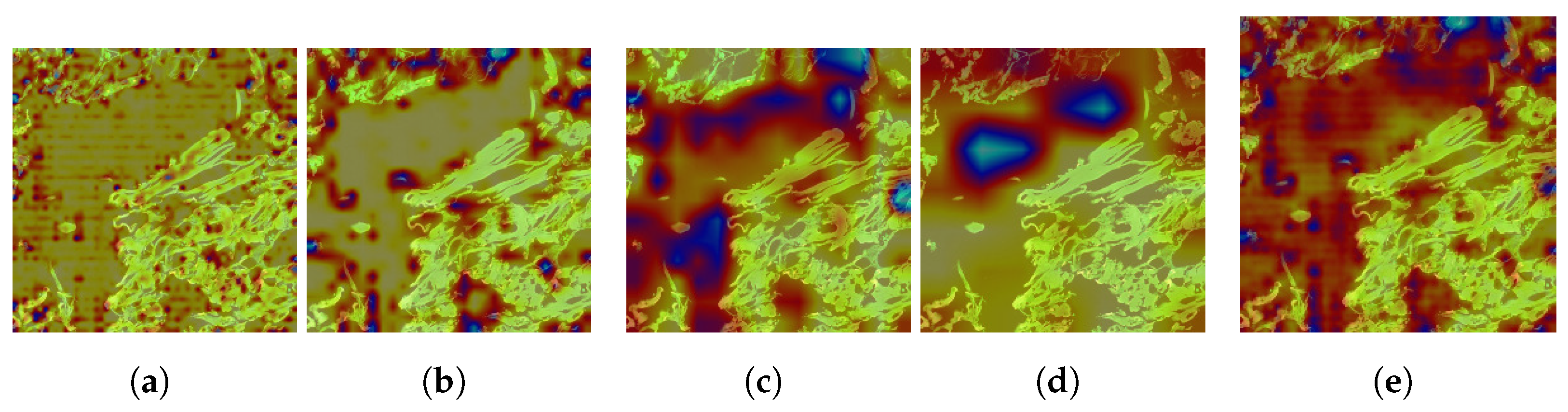
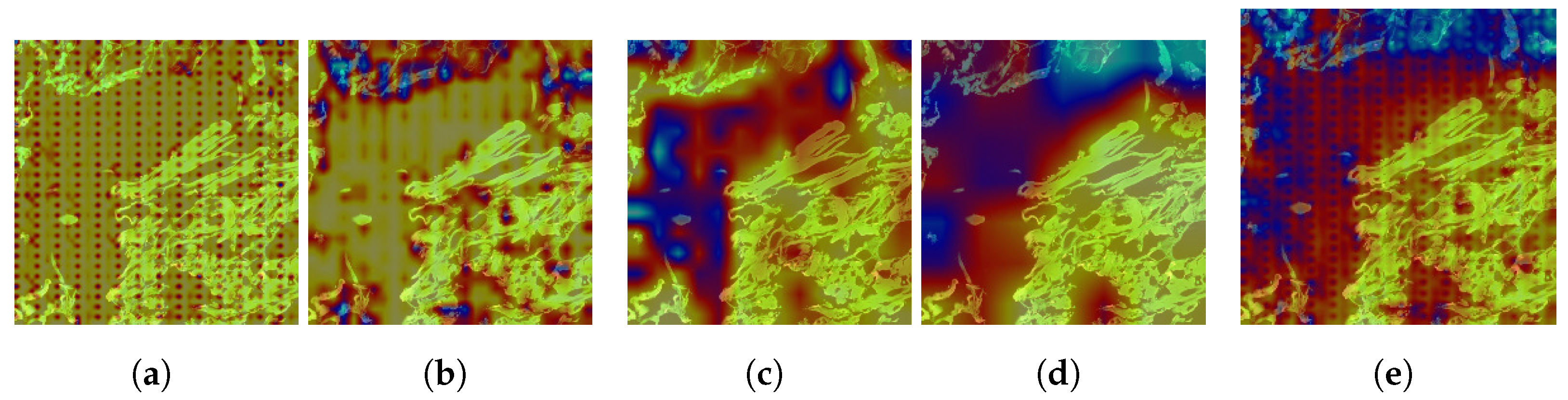
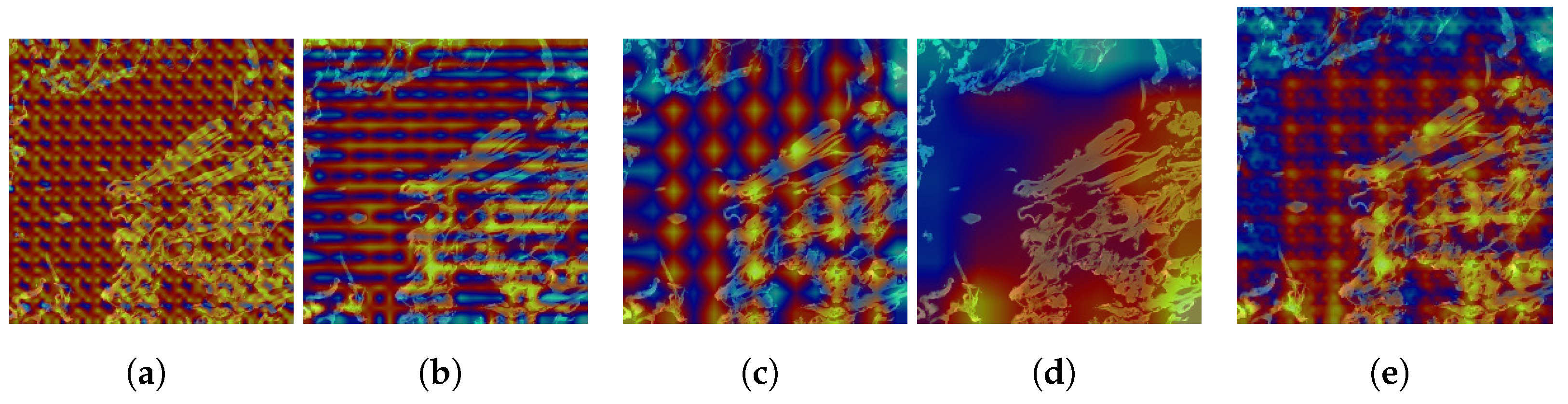
4.2.2. Explanation with CAM
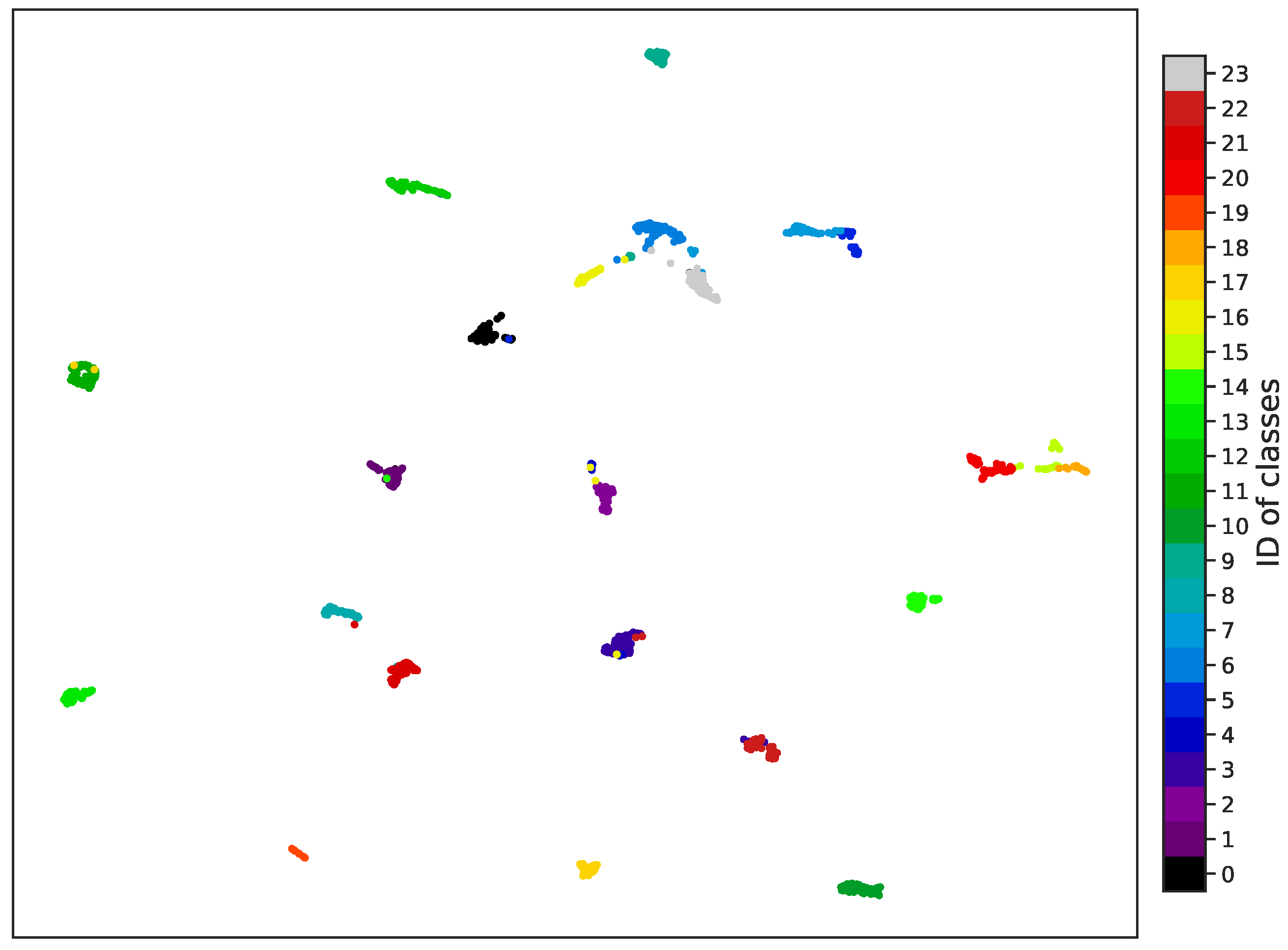
4.2.3. Visualization of Learned Embeddings
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Solar, M.; Castañeda, V.; Ñanculef, R.; Dombrovskaia, L.; Araya, M. A Data Ingestion Procedure towards a Medical Images Repository. Sensors 2024, 24, 4985. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, M.M.; Li, C.; Wu, X.; Yao, Y.; Hu, Z.; Jiang, T.; Li, X.; Qi, S. A Survey for Cervical Cytopathology Image Analysis Using Deep Learning. IEEE Access 2020, 8, 61687–61710. [Google Scholar] [CrossRef]
- Solar, M.; Aguirre, P. Deep learning techniques to process 3D chest CT. J. Univ. Comput. Sci. 2024, 30, 758. [Google Scholar] [CrossRef]
- Hegde, N.; Hipp, J.D.; Liu, Y.; Emmert-Buck, M.; Reif, E.; Smilkov, D.; Terry, M.; Cai, C.J.; Amin, M.B.; Mermel, C.H.; et al. Similar image search for histopathology: SMILY. Npj Digit. Med. 2019, 2, 56. [Google Scholar] [CrossRef]
- Hashimoto, N.; Takagi, Y.; Masuda, H.; Miyoshi, H.; Kohno, K.; Nagaishi, M.; Sato, K.; Takeuchi, M.; Furuta, T.; Kawamoto, K.; et al. Case-based similar image retrieval for weakly annotated large histopathological images of malignant lymphoma using deep metric learning. Med. Image Anal. 2023, 85, 102752. [Google Scholar] [CrossRef]
- Kumar, A.; Kim, J.; Cai, W.; Fulham, M.; Feng, D. Content-Based Medical Image Retrieval: A Survey of Applications to Multidimensional and Multimodality Data. J. Digit. Imaging 2013, 26, 1025–1039. [Google Scholar] [CrossRef]
- Abdelsamea, M.M.; Zidan, U.; Senousy, Z.; Gaber, M.M.; Rakha, E.; Ilyas, M. A survey on artificial intelligence in histopathology image analysis. Wiley Data Min. Knowl. Discov. 2022, 12, e1474. [Google Scholar] [CrossRef]
- Sikaroudi, M.; Hosseini, M.; Gonzalez, R.; Rahnamayan, S.; Tizhoosh, H.R. Generalization of vision pre-trained models for histopathology. Sci. Rep. 2023, 13, 6065. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Iqbal, S.; Qureshi, A.N.; Alhussein, M.; Choudhry, I.A.; Aurangzeb, K.; Khan, T.M. Fusion of Textural and Visual Information for Medical Image Modality Retrieval Using Deep Learning-Based Feature Engineering. IEEE Access 2023, 11, 93238–93253. [Google Scholar] [CrossRef]
- Deng, J.; Guo, J.; Liu, T.; Gong, M.; Zafeiriou, S. Sub-center ArcFace: Boosting Face Recognition by Large-Scale Noisy Web Faces. In Proceedings of the Computer Vision—ECCV 2020, Glasgow, UK, 23–28 August 2000; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 741–757. [Google Scholar]
- Shafiei, S.; Babaie, M.; Kalra, S.; Tizhoosh, H.R. Colored Kimia Path24 Dataset: Configurations and Benchmarks with Deep Embeddings. arXiv 2021, arXiv:2102.07611. [Google Scholar] [CrossRef]
- Ando, D.M.; McLean, C.Y.; Berndl, M. Improving Phenotypic Measurements in High-Content Imaging Screens. bioRxiv 2017. [Google Scholar] [CrossRef]
- Yang, P.; Zhai, Y.; Li, L.; Lv, H.; Wang, J.; Zhu, C.; Jiang, R. A deep metric learning approach for histopathological image retrieval. Methods 2020, 179, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Tabatabaei, Z.; Colomer, A.; Moll, J.O.; Naranjo, V. Toward More Transparent and Accurate Cancer Diagnosis With an Unsupervised CAE Approach. IEEE Access 2023, 11, 143387–143401. [Google Scholar] [CrossRef]
- Mohammad Alizadeh, S.; Sadegh Helfroush, M.; Müller, H. A novel Siamese deep hashing model for histopathology image retrieval. Expert Syst. Appl. 2023, 225, 120169. [Google Scholar] [CrossRef]
- Kather, J.N.; Weis, C.A.; Bianconi, F.; Melchers, S.M.; Schad, L.R.; Gaiser, T.; Marx, A.; Zöllner, F.G. Multi-class texture analysis in colorectal cancer histology. Sci. Rep. 2016, 6, 27988. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Tabatabaei, Z.; Colomer, A.; Moll, J.O.; Naranjo, V. Siamese Content-based Search Engine for a More Transparent Skin and Breast Cancer Diagnosis through Histological Imaging. arXiv 2024, arXiv:2401.08272. [Google Scholar] [CrossRef]
- Iqbal, S.; Qureshi, A.N. A Heteromorphous Deep CNN Framework for Medical Image Segmentation Using Local Binary Pattern. IEEE Access 2022, 10, 63466–63480. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Müller, H.; Zhang, S. Large-scale retrieval for medical image analytics: A comprehensive review. Med. Image Anal. 2018, 43, 66–84. [Google Scholar] [CrossRef]
- Yang, X.; Li, C.; He, R.; Yang, J.; Sun, H.; Jiang, T.; Grzegorzek, M.; Li, X.; Liu, C. CAISHI: A benchmark histopathological H&E image dataset for cervical adenocarcinoma in situ identification, retrieval and few-shot learning evaluation. Data Brief 2024, 53, 110141. [Google Scholar] [CrossRef]
- Tizhoosh, H.; Maleki, D.; Rahnamayan, S. Harmonizing the Scale: An End-to-End Self-Supervised Framework for Cross-Modal Search and Retrieval in Histopathology Archives. 2023. Available online: https://www.researchsquare.com/article/rs-3650733/v1 (accessed on 10 July 2024). [CrossRef]
- Shao, S.; Chen, K.; Karpur, A.; Cui, Q.; Araujo, A.; Cao, B. Global Features are All You Need for Image Retrieval and Reranking. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Los Alamitos, CA, USA, 1–6 October 2023; pp. 11002–11012. [Google Scholar] [CrossRef]
- Cao, B.; Araujo, A.; Sim, J. Unifying Deep Local and Global Features for Image Search. In Proceedings of the Computer Vision—ECCV 2020, Glasgow, UK, 23–28 August 2000; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 726–743. [Google Scholar]
- Zhang, Z.; Wang, L.; Zhou, L.; Koniusz, P. Learning Spatial-context-aware Global Visual Feature Representation for Instance Image Retrieval. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Los Alamitos, CA, USA, 2–3 October 2023; pp. 11216–11225. [Google Scholar] [CrossRef]
- Radenović, F.; Tolias, G.; Chum, O. Fine-Tuning CNN Image Retrieval with No Human Annotation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Rabinovich, A.; Berg, A.C. ParseNet: Looking Wider to See Better. arXiv 2015, arXiv:1506.04579. [Google Scholar] [CrossRef]
- Woo, S.; Debnath, S.; Hu, R.; Chen, X.; Liu, Z.; Kweon, I.S.; Xie, S. ConvNeXt V2: Co-designing and Scaling ConvNets with Masked Autoencoders. In Proceedings of the 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023; pp. 16133–16142. [Google Scholar] [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- Wang, H.; Wang, Y.; Zhou, Z.; Ji, X.; Gong, D.; Zhou, J.; Li, Z.; Liu, W. CosFace: Large Margin Cosine Loss for Deep Face Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 5265–5274. [Google Scholar] [CrossRef]
- Gamper, J.; Koohbanani, N.A.; Benet, K.; Khuram, A.; Rajpoot, N. PanNuke: An open pan-cancer histology dataset for nuclei instance segmentation and classification. In Proceedings of the European Congress on Digital Pathology, Warwick, UK, 10–13 April 2019; Springer: Cham, Switzerland, 2019; pp. 11–19. [Google Scholar]
- Gamper, J.; Koohbanani, N.A.; Graham, S.; Jahanifar, M.; Khurram, S.A.; Azam, A.; Hewitt, K.; Rajpoot, N. PanNuke Dataset Extension, Insights and Baselines. arXiv 2020, arXiv:2003.10778. [Google Scholar]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Babaie, M.; Kalra, S.; Sriram, A.; Mitcheltree, C.; Zhu, S.; Khatami, A.; Rahnamayan, S.; Tizhoosh, H.R. Classification and Retrieval of Digital Pathology Scans: A New Dataset. arXiv 2017, arXiv:1705.07522. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv 2019, arXiv:1912.01703. [Google Scholar] [CrossRef]
- Douze, M.; Guzhva, A.; Deng, C.; Johnson, J.; Szilvasy, G.; Mazaré, P.E.; Lomeli, M.; Hosseini, L.; Jégou, H. The Faiss library. arXiv 2024, arXiv:2401.08281. [Google Scholar] [CrossRef]
- Rosenthal, J.; Carelli, R.; Omar, M.; Brundage, D.; Halbert, E.; Nyman, J.; Hari, S.N.; Van Allen, E.M.; Marchionni, L.; Umeton, R.; et al. Building Tools for Machine Learning and Artificial Intelligence in Cancer Research: Best Practices and a Case Study with the PathML Toolkit for Computational Pathology. Mol. Cancer Res. 2022, 20, 202–206. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized Gradient-based Visual Explanations for Deep Convolutional Networks. arXiv 2017, arXiv:1710.11063. [Google Scholar] [CrossRef]
| Strategy | Pre-Training Data | |||
|---|---|---|---|---|
| A | IN-1K | |||
| B | IN-1K + PanNuke | |||
| C | IN-1K + PanNuke + Kimia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuñez-Fernández , C.; Farias , H.; Solar , M. Content-Based Histopathological Image Retrieval. Sensors 2025, 25, 1350. https://doi.org/10.3390/s25051350
Nuñez-Fernández C, Farias H, Solar M. Content-Based Histopathological Image Retrieval. Sensors. 2025; 25(5):1350. https://doi.org/10.3390/s25051350
Chicago/Turabian StyleNuñez-Fernández , Camilo, Humberto Farias , and Mauricio Solar . 2025. "Content-Based Histopathological Image Retrieval" Sensors 25, no. 5: 1350. https://doi.org/10.3390/s25051350
APA StyleNuñez-Fernández , C., Farias , H., & Solar , M. (2025). Content-Based Histopathological Image Retrieval. Sensors, 25(5), 1350. https://doi.org/10.3390/s25051350








