Method of Forearm Muscles 3D Modeling Using Robotic Ultrasound Scanning
Abstract
:1. Introduction
2. Materials and Methods
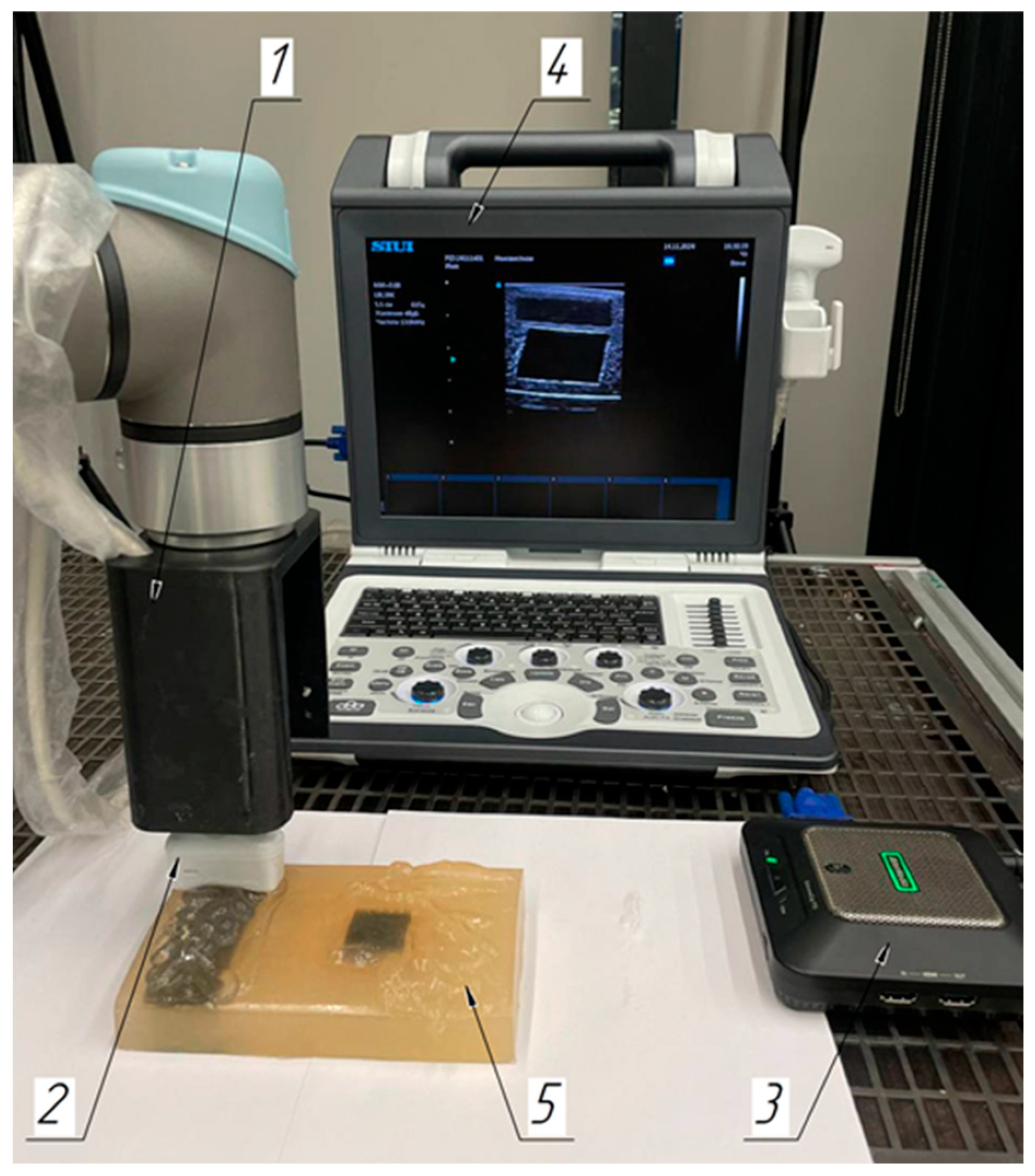
2.1. Laboratory Facilities Diagram
2.2. Subjects
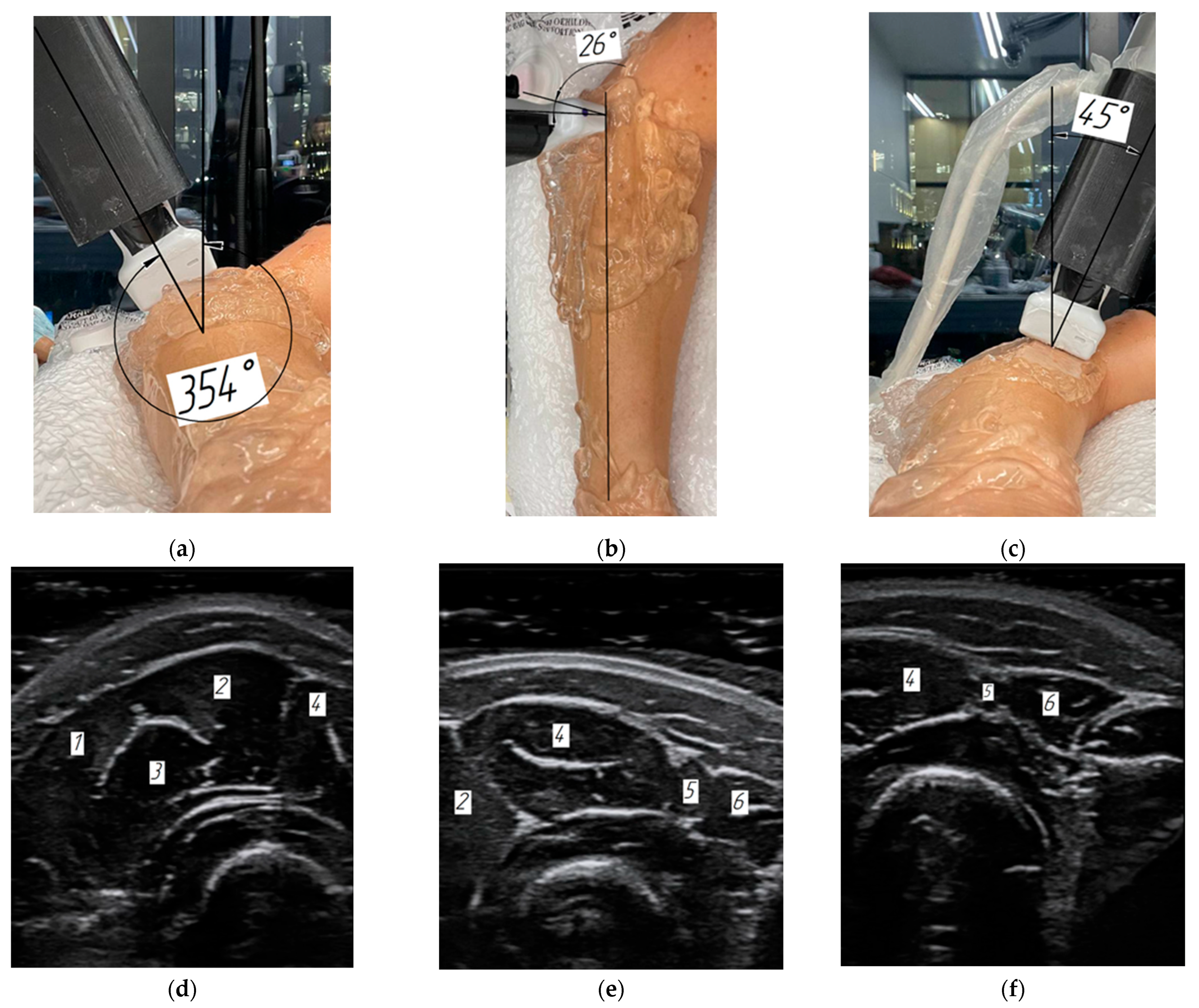
2.3. Robotic Scanning
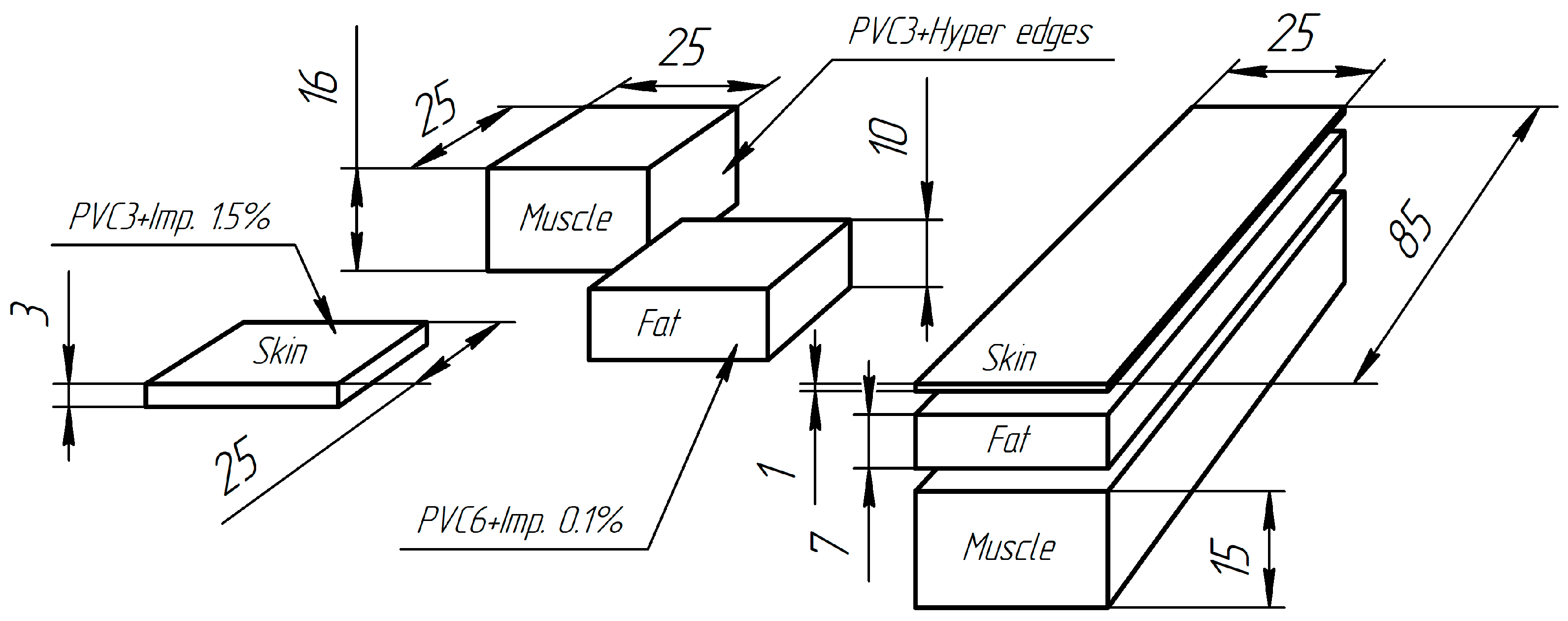
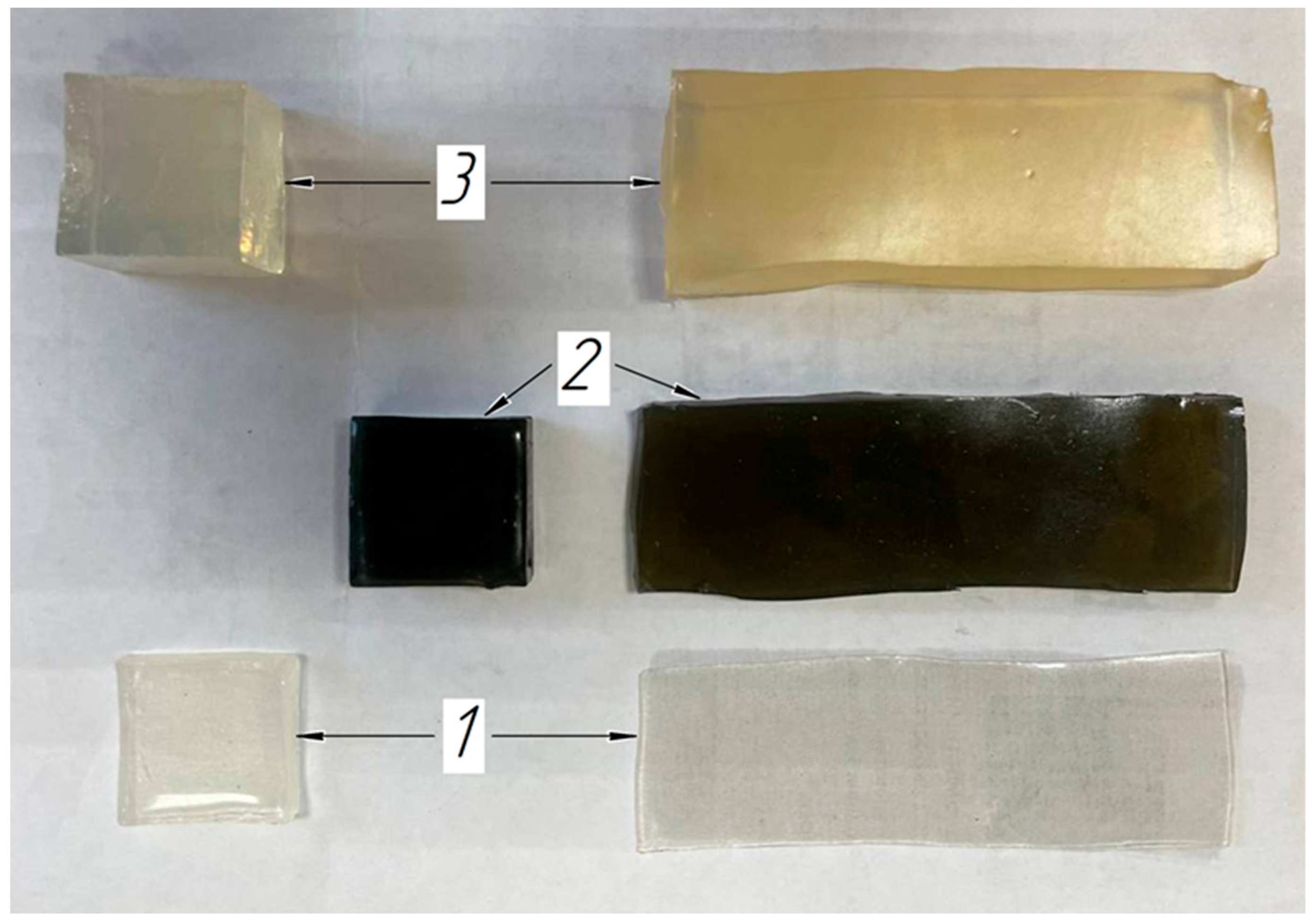
2.4. Soft Tissue Phantom Development
3. Results
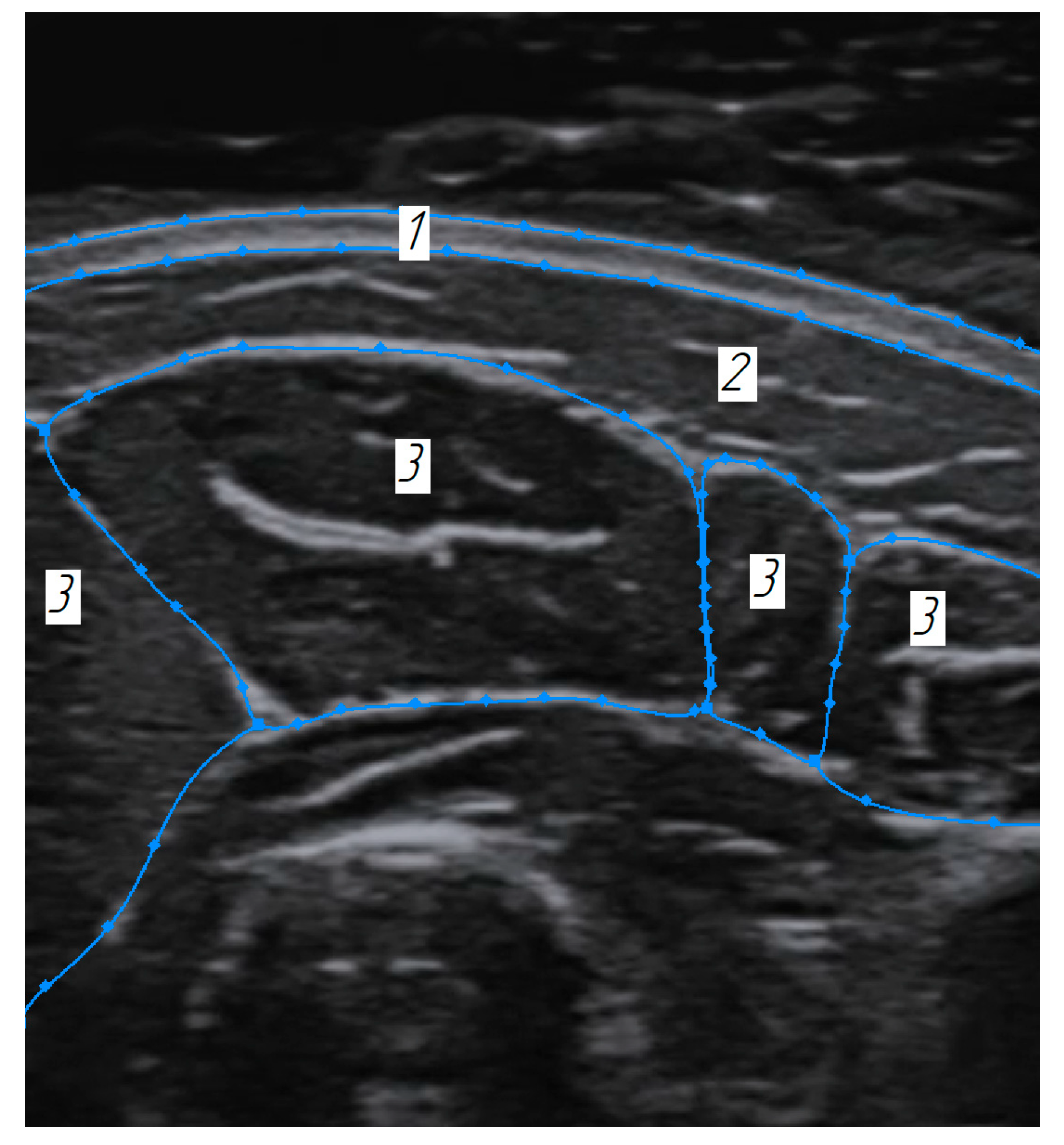
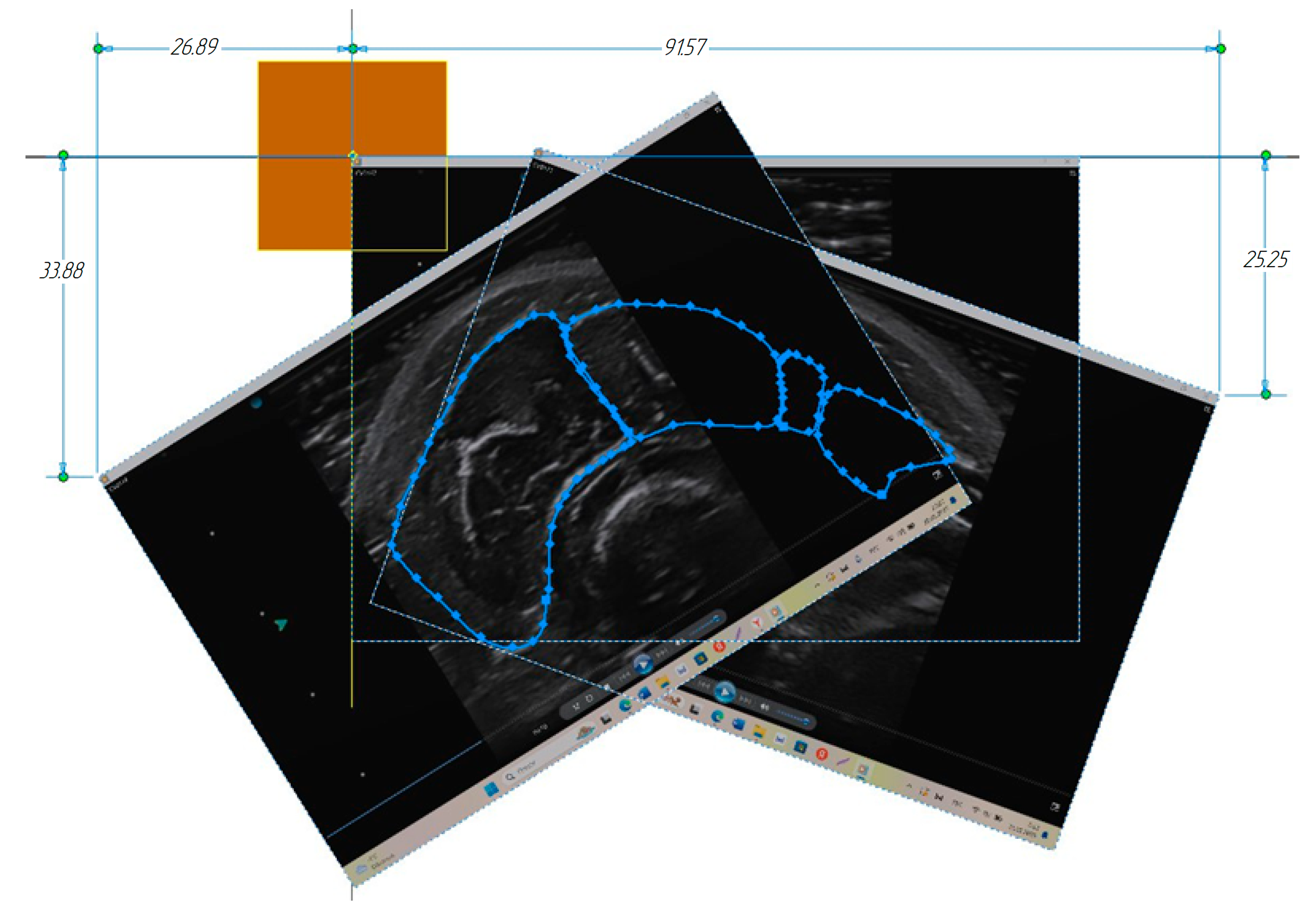
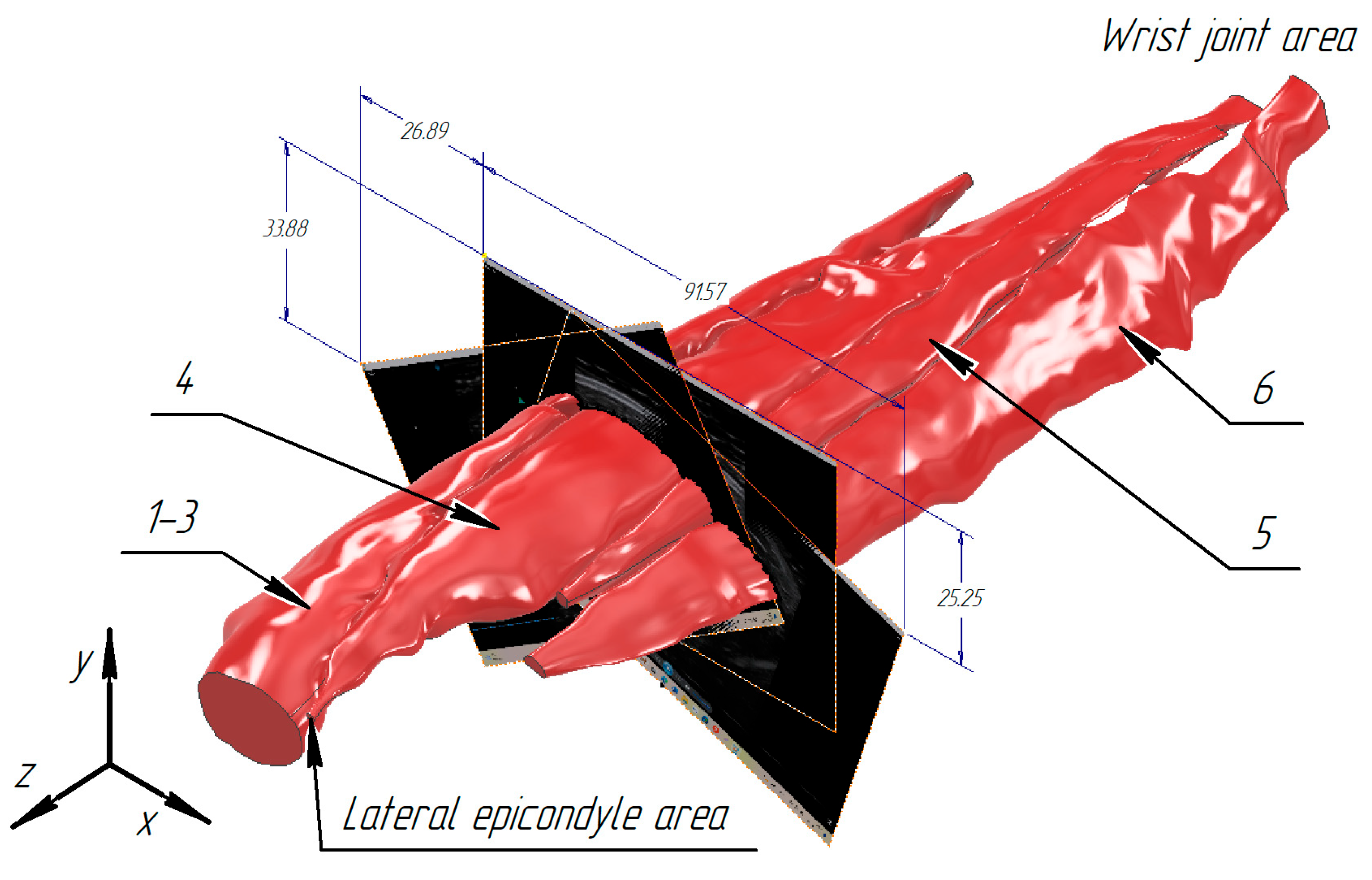
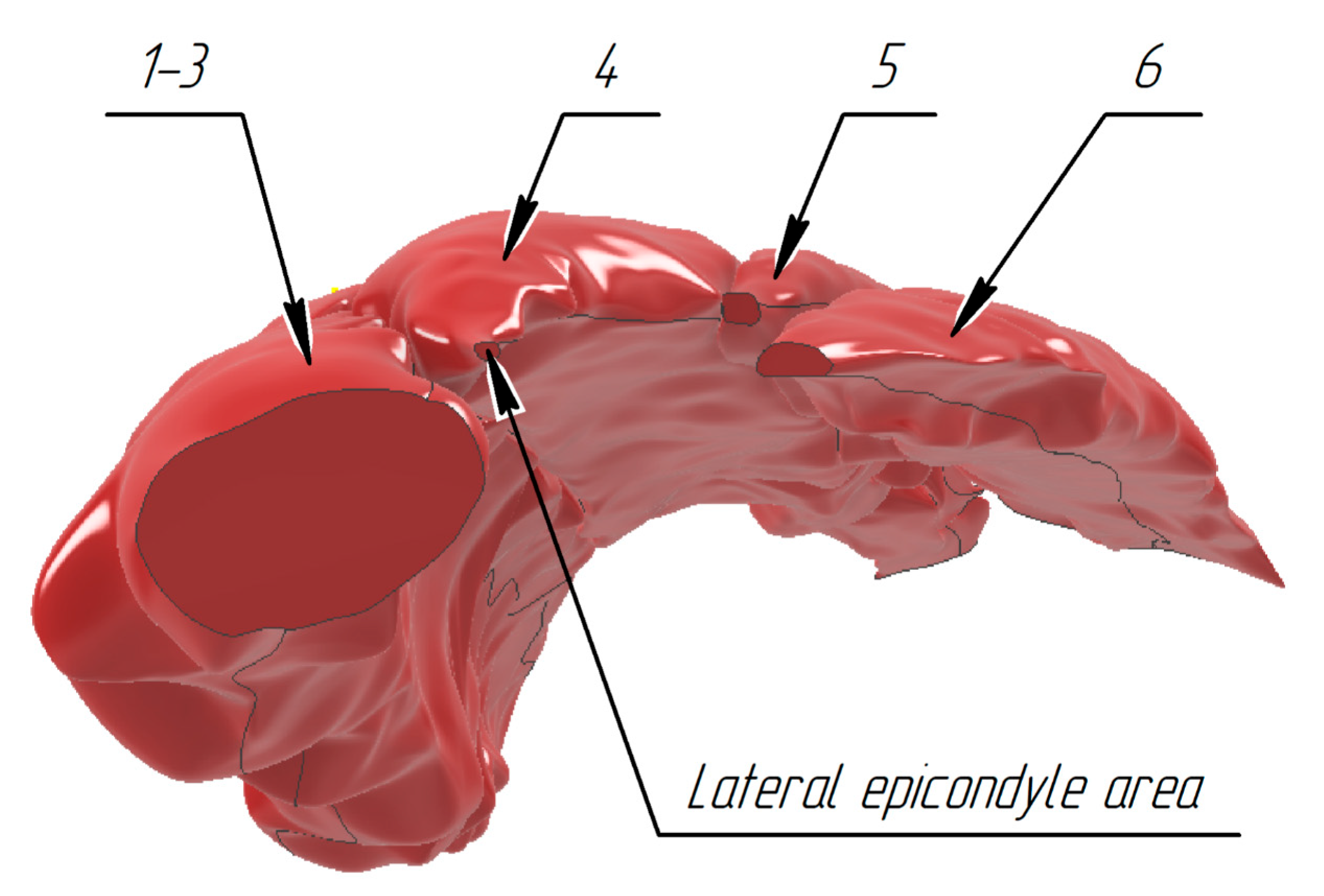
3.1. Construction of the Forearm Muscles’ Volumetric Models
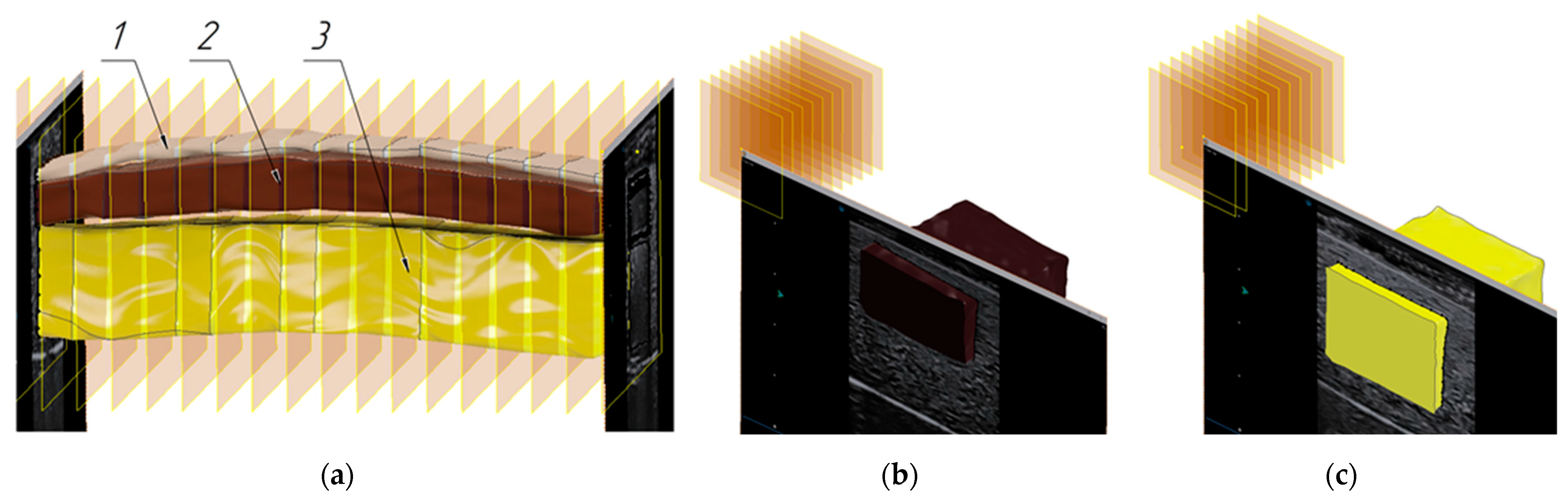
3.2. Verification of the Methodology for Constructing the Soft Tissue Volumetric Models
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
References
- Ortiz-Catalan, M.; Zbinden, J.; Millenaar, J.; D’Accolti, D.; Controzzi, M.; Clemente, F.; Cappello, L.; Earley, E.J.; Mastinu, E.; Kolankowska, J.; et al. A highly integrated bionic hand with neural control and feedback for use in daily life. Sci. Robot. 2023, 8, eadf7360. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lei, Y.; Shah, A.; Schein, G.; Ghaednia, H.; Schwab, J.; Harteveld, C.; Mueller, S. MuscleRehab: Improving unsupervised physical rehabilitation by monitoring and visualizing muscle engagement. In Proceedings of the 35th Annual ACM Symposium on User Interface Software and Technology, Bend, OR, USA, 29 October–2 November 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 1–14. [Google Scholar] [CrossRef]
- Murphy, E.K.; Skinner, J.; Martucci, M.; Rutkove, S.B.; Halter, R.J. Toward electrical impedance tomography coupled ultrasound imaging for assessing muscle health. IEEE Trans. Med. Imaging 2018, 38, 1409–1419. [Google Scholar] [PubMed]
- Briko, A.N.; Selutina, S.E.; Parnovskaya, A.D.; Emelin, M.E. Determination of Tissue Properties Based on Modeling and Electrical Impedance Registration. In Proceedings of the 2020 Ural Symposium on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 14–15 May 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 0028–0031. [Google Scholar]
- Kapravchuk, V.; Briko, A.; Kobelev, A.; Hammoud, A.; Shchukin, S. An Approach to using electrical impedance myography signal sensors to assess morphofunctional changes in tissue during muscle contraction. Biosensors 2024, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Youssef Baby, L.; Bedran, R.S.; Doumit, A.; El Hassan, R.H.; Maalouf, N. Past, present, and future of electrical impedance tomography and myography for medical applications: A scoping review. Front. Bioeng. Biotechnol. 2024, 12, 1486789. [Google Scholar] [CrossRef]
- Pandeya, S.R.; Nagy, J.A.; Riveros, D.; Semple, C.; Taylor, R.S.; Sanchez, B.; Rutkove, S.B. Relationships between in vivo surface and ex vivo electrical impedance myography measurements in three different neuromuscular disorder mouse models. PLoS ONE 2021, 16, e0259071. [Google Scholar]
- Hu, H.; Lo, W.L.A.; Wang, X.; Li, L.; Zhou, P. Interrater and intrarater reliability of electrical impedance myography: A comparison between large and small handheld electrode arrays. J. Healthc. Eng. 2021, 2021, 1–8. [Google Scholar] [CrossRef]
- Shcherbachev, A.; Alexandrov, A.; Kudashov, I.; Apolikhin, O.; Savostyanov, V.; Pavlov, A. Development of Multilayer Impedance Sensors for Dynamic Monitoring of Respiratory Rate. In Proceedings of the 2023 IEEE Ural-Siberian Conference on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 15–17 May 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 016–019. [Google Scholar]
- Mugeb, A.; Belsheva, M.; Smirnova, A. Influence of Arterial Blood Flow on Impedance Plethysmography. In Proceedings of the 2022 Ural-Siberian Conference on Computational Technologies in Cognitive Science, Genomics and Biomedicine (CSGB), Novosibirsk, Russia, 4–8 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 140–145. [Google Scholar]
- Shcherbachev, A.; Kudashov, I.; Itkin, G.; Bychkov, E.; Pavlov, A.; Govorin, A. Approximation Model for Electroimpedance Measurement Finite Diastolic Volume AHV. In Proceedings of the 2022 Ural-Siberian Conference on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT), Yekaterinburg, Russia, 19–21 September 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 055–058. [Google Scholar]
- Buchanan, T.S.; Lloyd, D.G.; Manal, K.; Besier, T.F. Neuromusculoskeletal modeling: Estimation of muscle forces and joint moments and movements from measurements of neural command. J. Appl. Biomech. 2024, 20, 367–395. [Google Scholar]
- Saini, H.; Klotz, T.; Röhrle, O. Modelling motor units in 3D: Influence on muscle contraction and joint force via a proof of concept simulation. Biomech. Model Mechanobiol 2023, 22, 593–610. [Google Scholar] [CrossRef]
- Berranen, Y.; Hayashibe, M.; Gilles, B.; Guiraud, D. 3D volumetric muscle modeling for real-time deformation analysis with FEM. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 4863–4866. [Google Scholar]
- Dereshgı, H.A.; Serbest, K.; Şahin, S.N.; Balık, B. Skeletal muscle mechanics from hill-based muscle model to computer applications: State of the art review. J. Smart Syst. Res. 2021, 2, 27–39. [Google Scholar]
- Komaritzan, M.; Wenninger, S.; Botsch, M. Inside humans: Creating a simple layered anatomical model from human surface scans. Front. Virtual Real. 2021, 2, 694244. [Google Scholar]
- Andreassen, T.E.; Hume, D.R.; Hamilton, L.D.; Walker, K.E.; Higinbotham, S.E.; Shelburne, K.B. Three dimensional lower extremity musculoskeletal geometry of the visible human female and male. Sci. Data 2023, 10, 34. [Google Scholar] [PubMed]
- Wang, Z.; Destro, A.; Petersson, S.; Cenni, F.; Wang, R. In vivo 3D muscle architecture quantification based on 3D freehand ultrasound and magnetic resonance imaging. J. Biomech. 2023, 152, 111567. [Google Scholar]
- Blemker, S.S. In vivo imaging of skeletal muscle form and function: 50 years of insight. J. Biomech. 2023, 158, 111745. [Google Scholar] [CrossRef]
- Berry, D.B.; Gordon, J.A.; Adair, V.; Frank, L.R.; Ward, S.R. From Voxels to Physiology: A Review of Diffusion Magnetic Resonance Imaging Applications in Skeletal Muscle. Magn. Reson. Imaging 2025, 61, 595–615. [Google Scholar] [CrossRef]
- Kwak, Y.; Kim, J.; Lee, K.M.; Koo, S. Increase of stiffness in plantar fat tissue in diabetic patients. J. Biomech. 2020, 107, 109857. [Google Scholar] [CrossRef]
- Bachasson, D.; Ayaz, A.C.; Mosso, J.; Canal, A.; Boisserie, J.; Araujo, E.C.; Benveniste, O.; Reyngoudt, H.; Marty, B.; Carlier, P.G.; et al. Lean regional muscle volume estimates using explanatory bioelectrical models in healthy subjects and patients with muscle wasting. J. Cachexia Sarcopenia Muscle 2021, 12, 39–51. [Google Scholar] [CrossRef]
- Anderson, D.E.; Bean, J.F.; Holt, N.E.; Keel, J.C.; Bouxsein, M.L. Computed tomography-based muscle attenuation and electrical impedance myography as indicators of trunk muscle strength independent of muscle size in older adults. Am. J. Phys. Med. Rehabil. 2014, 93, 553–561. [Google Scholar] [CrossRef]
- Abellan van Kan, G.; Cedarbaum, J.M.; Cesari, M.; Dahinden, P.; Fariello, R.G.; Fielding, R.A.; Goodpaster, B.H.; Hettwer, S.; Isaac, M.; Laurent, D.; et al. Sarcopenia: Biomarkers and imaging (International Conference on Sarcopenia research). J. Nutr. Health Aging 2011, 15, 834–846. [Google Scholar]
- Salminger, S.; Cedarbaum, J.M.; Cesari, M.; Dahinden, P.; Fariello, R.G.; Fielding, R.A.; Goodpaster, B.H.; Hettwer, S.; Isaac, M.; Laurent, D.; et al. Current rates of prosthetic usage in upper-limb amputees–have innovations had an impact on device acceptance? Disabil. Rehabil. 2022, 44, 3708–3713. [Google Scholar] [CrossRef]
- Sanders, J.E.; Fatone, S. Residual limb volume change: Systematic review of measurement and management. J. Rehabil. Res. Dev. 2011, 48, 949. [Google Scholar] [CrossRef]
- Briko, A.N.; Parnovskaya, A.D.; Kapravchuk, V.V.; Dyachenkova, S.I.; Shchukin, S.I.; Kobelev, A.V.; Vasiliev, Y.A.; Petryaikin, A.V. Parametric assessment of morphological changes in forearm tissues when performing basic actions with a hand based on MRI: Methodological aspects. Biomed. Radioelekronika Biomed. Radioeng. 2021, 24, 6–16. [Google Scholar]
- MacGillivray, T.J.; Ross, E.; Simpson, H.A.; Greig, C.A. 3D freehand ultrasound for in vivo determination of human skeletal muscle volume. Ultrasound Med. Biol. 2009, 35, 928–935. [Google Scholar] [PubMed]
- Chen, E.C.; Groves, L.A.; Nisar, H.; Carnahan, P.K.; Cambranis-Romero, J.; Peters, T.M. Freehand 3D ultrasound: Principle and clinical applications. In 3D Ultrasound; CRC Press: Boca Raton, FL, USA, 2023; pp. 35–48. [Google Scholar]
- Zwiebel, W.J.; Pellerito, J.S. Introduction to Vascular Ultrasonography; Elsevier Saunders: Philadelphia, PA, USA, 2005; pp. 19–89. [Google Scholar]
- Jiang, Z.; Salcudean, S.E.; Navab, N. Robotic ultrasound imaging: State-of-the-art and future perspectives. Med. Image Anal. 2023, 89, 102878. [Google Scholar] [PubMed]
- Wu, G.; Shen, X.; Serebrenny, V. Attention Guided In-hand Mechanical Tools Recognition in Human-Robot Collaborative Process. In Proceedings of the International Conference on Interactive Collaborative Robotics, Baku, Azerbaijan, 25–29 October 2023; Springer Nature: Cham, Switzerland, 2023; pp. 1–12. [Google Scholar]
- Filko, D.; Nyarko, E.K. 2D/3D wound segmentation and measurement based on a robot-driven reconstruction system. Sensors 2023, 23, 3298. [Google Scholar] [CrossRef]
- Yuschenko, A. Interactive Collaborative Robotics–New Results and New Problems. In Communications in Computer and Information Science, Proceedings of the Modern Problems of Robotics: Second International Conference MPoR, Moscow, Russia, 25–26 March 2020; Springer International Publishing: New York, NY, USA, 2020; pp. 29–43. [Google Scholar]
- Adriaans, C.A.; Wijkhuizen, M.; van Karnenbeek, L.M.; Geldof, F.; Dashtbozorg, B. Trackerless 3D freehand ultrasound reconstruction: A review. Appl. Sci. 2024, 14, 7991. [Google Scholar] [CrossRef]
- Dinescu, S.C. Applications of artificial intelligence in musculoskeletal ultrasound: Narrative review. Front. Med. 2023, 10, 1286085. [Google Scholar]
- Huang, Q.; Zhou, J.; Li, Z. Review of robot-assisted medical ultrasound imaging systems: Technology and clinical applications. Neurocomputing 2023, 559, 126790. [Google Scholar]
- Grushko, S.; Spurnỳ, T.; Černỳ, M. Control methods for transradial prostheses based on remnant muscle activity and its relationship with proprioceptive feedback. Sensors 2020, 20, 4883. [Google Scholar] [CrossRef]
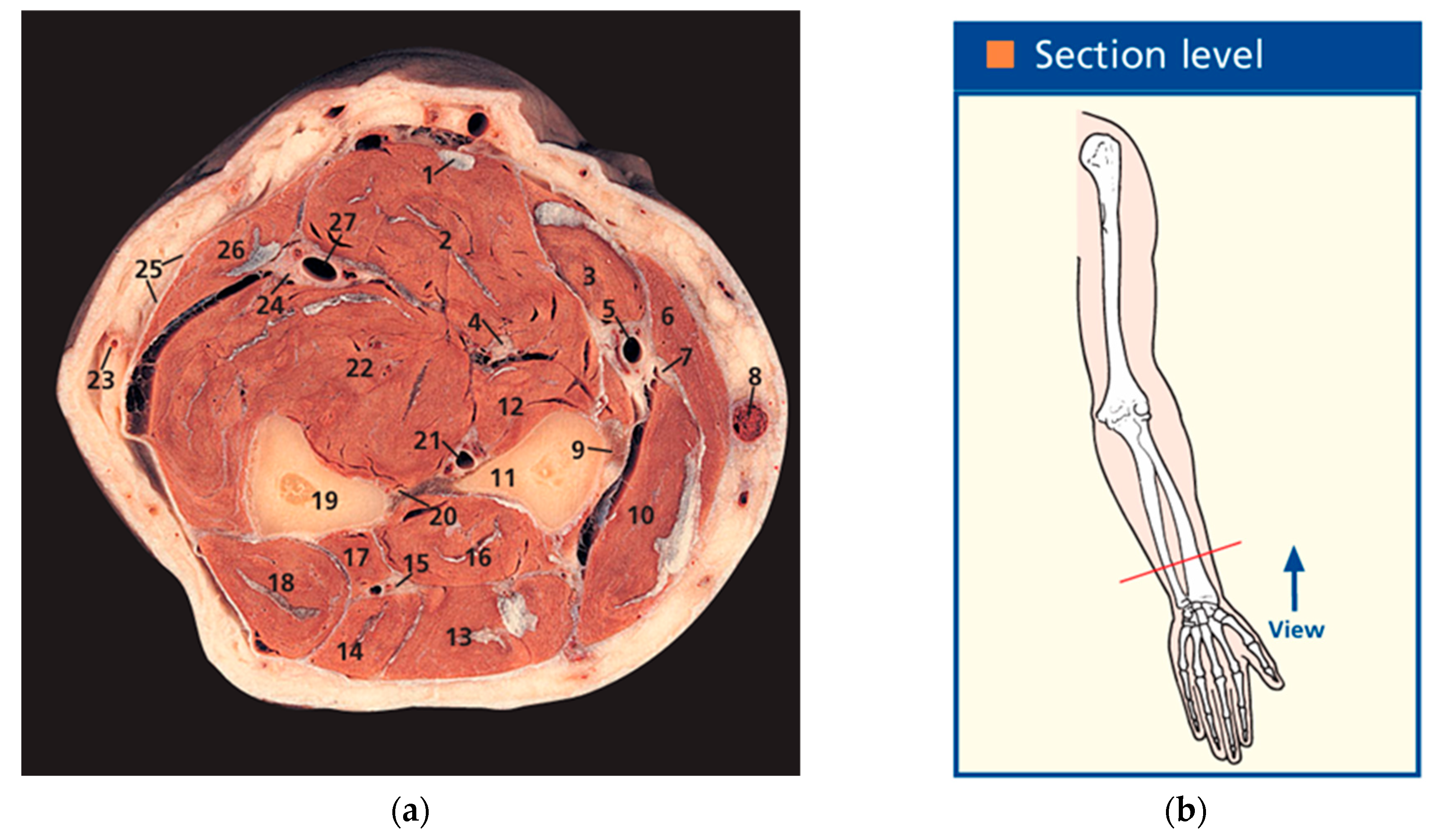
- Ellis, H.; Logan, B.M.; Dixon, A.K. Human Sectional Anatomy: Atlas of Body Sections, CT and MRI Images, 3rd ed.; Hodder Arnold: London, UK, 2010; Volume 92, pp. 178–179. [Google Scholar] [CrossRef]
- Jawli, A.; Aldehani, W.; Nabi, G.; Huang, Z. Tissue-mimicking material fabrication and properties for multiparametric ultrasound phantoms: A systematic review. Bioengineering 2024, 11, 620. [Google Scholar] [CrossRef]
- Groves, L.A.; Keita, M.; Talla, S.; Kikinis, R.; Fichtinger, G.; Mousavi, P.; Camara, M. A review of low-cost ultrasound compatible phantoms. IEEE Trans. Biomed. Eng. 2023, 70, 3436–3448. [Google Scholar]
- Leonov, D.; Venidiktova, D.; Costa-Júnior, J.F.S.; Nasibullina, A.; Tarasova, O.; Pashinceva, K.; Vetsheva, N.; Bulgakova, J.; Kulberg, N.; Borsukov, A.; et al. Development of an anatomical breast phantom from polyvinyl chloride plastisol with lesions of various shape, elasticity and echogenicity for teaching ultrasound examination. Int. J. Comput. Assist. Radiol. Surg. 2024, 19, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Curiel, L.; Chopra, R.; Hynynen, K. Progress in multimodality imaging: Truly simultaneous ultrasound and magnetic resonance imaging. IEEE Trans. Med. Imaging 2007, 26, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W. Fusion imaging of real-time ultrasonography with CT or MRI for hepatic intervention. Ultrasonography 2014, 33, 227. [Google Scholar] [CrossRef] [PubMed]
- D’Addazio, G.; Xhajanka, E.; Traini, T.; Santilli, M.; Rexhepi, I.; Murmura, G.; Caputi, S.; Sinjari, B. Accuracy of DICOM–DICOM vs. DICOM–STL Protocols in Computer-Guided Surgery: A Human Clinical Study. J. Clin. Med. 2022, 11, 2336. [Google Scholar] [CrossRef]
- Kamio, T.; Suzuki, M.; Asaumi, R.; Kawai, T. DICOM segmentation and STL creation for 3D printing: A process and software package comparison for osseous anatomy. 3D Print. Med. 2020, 6, 1–12. [Google Scholar] [CrossRef]
- Schrunder, A.F.; Rodriguez, S.; Rusu, A. A finite element analysis and circuit modelling methodology for studying electrical impedance myography of human limbs. IEEE Trans. Biomed. Eng. 2021, 69, 244–255. [Google Scholar] [CrossRef]
- Wan, J.N.; Zhou, H.Y.; Gao, Y.M.; Yang, J.J.; Vasić, Ž.L.; Cifrek, M.; Du, M. Optimization of the electrode configuration of electrical impedance myography for wearable application. Automatika 2020, 61, 475–481. [Google Scholar] [CrossRef]
- Katakis, S.; Barotsis, N.; Kakotaritis, A.; Tsiganos, P.; Economou, G.; Panagiotopoulos, E.; Panayiotakis, G. Muscle cross-sectional area segmentation in transverse ultrasound images using vision transformers. Diagnostics 2023, 13, 217. [Google Scholar] [CrossRef]








| Characteristic | Value |
|---|---|
| Collaborative 6-degree robotic manipulator Universal Robots UR5e | |
| Maximum working speed of the tool displacement | 1 m/s |
| Linear displacement speed | Not less than 1000 mm/s |
| Number of degrees of freedom | 6 |
| Positioning accuracy | ±0.1 mm |
| Manipulator position coordinates measurement range | 100 ± 30 mm |
| Force sensor registered values range along the x-y-z axes | 50.0 N |
| Torque moment registered range along the x-y-z axes | 10.0 N*m |
| Force sensor registered values accuracy along the x-y-z axes | 0.2 N |
| Torque moment registered accuracy along the x-y-z axes | 0.2 N*m |
| Ultrasound diagnostics system SIUI Apogee 1100 | |
| Ultrasound sensor type | Linear |
| Linear sensor frequency | 13 MHz |
| Maximum penetration depth | 300 mm |
| Number of greyscale gradations | 256 |
| Video capture card AverMedia ExtremeCap 910 | |
| Interface | USB 2.0 |
| Video in/video out | Digital: HDMI Analog: D-Sub |
| Video capture standards | In: to 1920 × 1080 60 r, Capture: to 1920 × 1080 30 r 10 Mbit/s |
| Model Name | Volume Value (with Relative Error) | Center of Gravity Along the | ||
|---|---|---|---|---|
| X-axis | Y-axis | Z-axis | ||
| Combined model of m. brachioradialis, m. extensors carpi radiales longus, m. extensors carpi radiales brevis | 52,469.046 mm3 (0.564647%) | 15.615 mm | −33.547 mm | −71.380 mm |
| m. extensor digitorum | 26,912.710 mm3 (0.883477%) | 32.978 mm | −22.674 mm | 94.406 mm |
| m. extensor digiti minimi | 6211.816 mm3 (0.876173%) | 43.686 mm | −22.801 mm | −117.131 mm |
| m. extensor carpi ulnaris | 15,123.671 mm3 (0.568765%) | 52.925 mm | −28.040 mm | −121.890 mm |
| General model | 100,717.243 mm3 (0.980515%) | 27.540 mm | −29.125 mm | −88.361 mm |
| Model Name | Volume Value (with Relative Error) | Areas of the Side Faces | Perimeters of the Side Faces | ||
|---|---|---|---|---|---|
| Imitation of the muscle layer (elongated shape) | 35,408.850 mm3 (0.209156%) | 398.64 mm2 | 334.99 mm2 | 81.31 mm | 74.24 mm |
| Imitation of the fat layer (elongated shape) | 17,979.904 mm3 (0.424175%) | 174.86 mm2 | 160.84 mm2 | 69.59 mm | 61.56 mm |
| Imitation of the muscle layer (square shape) | 7637.408 mm3 (0.059108%) | 310.21 mm2 | 299.02 mm2 | 74.59 mm | 73.15 mm |
| Imitation of the fat layer (square shape) | 10,556.336 mm3 (0.603168%) | 429.42 mm2 | 462.62 mm2 | 84.63 mm | 86.92 mm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapravchuk, V.; Ishkildin, A.; Briko, A.; Borde, A.; Kodenko, M.; Nasibullina, A.; Shchukin, S. Method of Forearm Muscles 3D Modeling Using Robotic Ultrasound Scanning. Sensors 2025, 25, 2298. https://doi.org/10.3390/s25072298
Kapravchuk V, Ishkildin A, Briko A, Borde A, Kodenko M, Nasibullina A, Shchukin S. Method of Forearm Muscles 3D Modeling Using Robotic Ultrasound Scanning. Sensors. 2025; 25(7):2298. https://doi.org/10.3390/s25072298
Chicago/Turabian StyleKapravchuk, Vladislava, Albert Ishkildin, Andrey Briko, Anna Borde, Maria Kodenko, Anastasia Nasibullina, and Sergey Shchukin. 2025. "Method of Forearm Muscles 3D Modeling Using Robotic Ultrasound Scanning" Sensors 25, no. 7: 2298. https://doi.org/10.3390/s25072298
APA StyleKapravchuk, V., Ishkildin, A., Briko, A., Borde, A., Kodenko, M., Nasibullina, A., & Shchukin, S. (2025). Method of Forearm Muscles 3D Modeling Using Robotic Ultrasound Scanning. Sensors, 25(7), 2298. https://doi.org/10.3390/s25072298







