Antibody-Based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins
Abstract
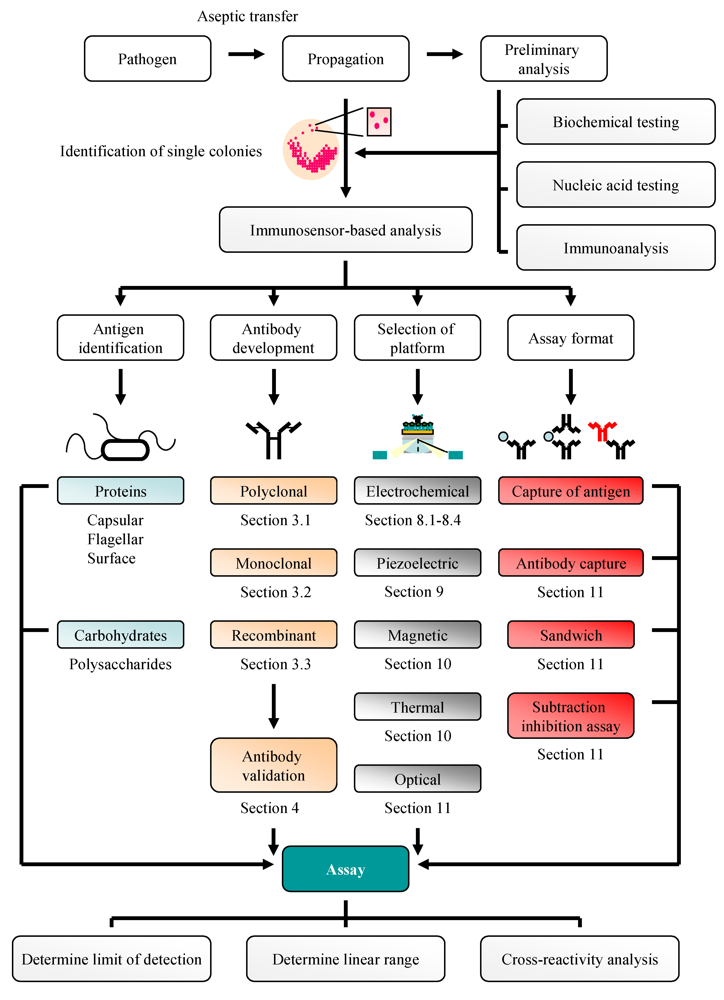
:1. Introduction
2. Bacteriological and Nucleic Acid-Based Analysis of Pathogenic Bacteria: A Traditional Approach
3. Antibodies: Production and Purification
3.1. Polyclonal Antibodies
3.2. Monoclonal Antibodies
3.3. Recombinant Antibodies
4. Antibody Selection
5. Use of Antibodies for Antigen Isolation/Enrichment Prior to Analysis
6. ELISA and Microarray-Based Pathogen Detection
7. Biosensors
8. Electrochemical Immunosensors
8.1. Amperometric Platforms
8.2. Impedimetric Platforms
8.3. Potentiometric Platforms
8.4. Conductimetric Platforms
9. Mass-Based Immunosensors
10. Thermometric and Magnetic Immunosensors
11. Optical Immunosensors
12. Immunosensor-Based Assays for the Detection of Other Bacterial Pathogens
13. Immunosensors for Fungal Pathogens and Mycotoxins
14. Immunosensor Assays for the Detection of Viral Pathogens, Marine Toxins and Parasites
15. Antibody-Based Biosensors: Potential Issues
16. Alternative Sensor-Based Platforms for Pathogen Detection
17. Conclusions
Acknowledgments
References and Notes
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar]
- Bhunia, A.K. Biosensors and bio-based methods for the separation and detection of foodborne pathogens. Adv. Food Nutr. Res. 2008, 54, 1–44. [Google Scholar]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O'Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Tech. 2003, 32, 3–13. [Google Scholar]
- Available online: www.oxoid.com/UK/blue/orgbrowse/orgbrowse.asp, Accession date: May 6, 2009.
- Kim, H.; Bhunia, A.K. SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7 and Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4853–4866. [Google Scholar]
- Hearty, S.; Leonard, P.; Quinn, J.; O'Kennedy, R. Production, characterisation and potential application of novel monoclonal antibody for rapid identification of virulent Listeria monocytogenes. J. Microbiol. Methods 2006, 66, 294–312. [Google Scholar]
- De Boer, E.; Beumer, R.R. Methodology for detection and typing of foodborne microorganisms. Int. J. Food Microbiol. 1999, 50, 119–130. [Google Scholar]
- O'Grady, J.; Ruttledge, M.; Sedano-Balbás, S.; Smith, T.J.; Barry, T.; Maher, M. Rapid detection of Listeria monocytogenes in food using culture enrichment combined with real-time PCR. Food Microbiol 2009, 26, 4–7. [Google Scholar]
- Brooks, B.W.; Devenish, J.; Lutze-Wallace, C.L.; Milnes, D.; Robertson, R.H.; Berlie-Surujballi, G. Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for the detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Vet. Microbiol. 2004, 103, 77–84. [Google Scholar]
- Dreux, N.; Albagnac, C.; Federighi, M.; Carlin, F.; Morris, C.E.; Nguyen-The, C. Viable but non-culturable Listeria monocytogenes on parsley leaves and absence of recovery to a culturable state. J. Appl. Microbiol. 2007, 103, 1272–1281. [Google Scholar]
- Asakura, H.; Panutdaporn, N.; Kawamoto, K.; Igimi, S.; Yamamoto, S.; Makino, S. Proteomic characterization of enterohemorrhagic Escherichia coli O157:H7 in the oxidation-induced viable but non-culturable state. Microbiol. Immunol. 2007, 51, 875–881. [Google Scholar]
- Uyttendaele, M.; Schukkink, R.; van Gemen, B.; Debevere, J. Development of NASBA, a nucleic acid amplification system, for identification of Listeria monocytogenes and comparison to ELISA and a modified FDA method. Int. J. Food Microbiol. 1995, 27, 77–89. [Google Scholar]
- Nadal, A.; Coll, A.; Cook, N.; Pla, M. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: Role of target mRNA secondary structure on NASBA design. J. Microbiol. Methods 2007, 68, 623–632. [Google Scholar]
- Toze, S. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 1999, 33, 3545–3556. [Google Scholar]
- Rodríguez-Lázaro, D.; D'Agostino, M.; Herrewegh, A.; Pla, M.; Cook, N.; Ikonomopoulos, J. Real-time PCR-based methods for detection of Mycobacterium avium subsp. Paratuberculosis in water and milk. Int. J. Food Microbiol. 2005, 101, 93–104. [Google Scholar]
- Fu, Z.; Rogelj, S.; Kieft, T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005, 99, 47–57. [Google Scholar]
- Renwick, L.; Hardie, A.; Girvan, E.K.; Smith, M.; Leadbetter, G.; Claas, E.; Morrison, D.; Gibb, A.P.; Dave, J.; Templeton, K.E. Detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine leukocidin directly from clinical samples and the development of a multiplex assay using real-time polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 791–796. [Google Scholar]
- Rodríguez-Lázaro, D.; Jofré, A.; Aymerich, T.; Hugas, M.; Pla, M. Rapid quantitative detection of Listeria monocytogenes in meat products by real-time PCR. Appl. Environ. Microbiol. 2004, 70, 6299–6301. [Google Scholar]
- Park, H.J.; Kim, H.J.; Park, S.H.; Shin, E.G.; Kim, J.H.; Kim, H.Y. Direct and quantitative analysis of Salmonella enterica serovar Typhimurium using real-time PCR from artificially contaminated chicken meat. J. Microbiol. Biotechnol. 2008, 18, 1453–1458. [Google Scholar]
- Wang, L.; Li, Y.; Mustaphai, A. Rapid and simultaneous quantitation of Escherichia coli O157:H7, Salmonella, and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J. Food Prot. 2007, 70, 1366–1372. [Google Scholar]
- Jofré, A.; Martin, B.; Garriga, M.; Hugas, M.; Pla, D.; Rodríguez-Lázaro, D.; Aymerich, T. Simultaneous detection of Listeria monocytogenes and Salmonella by multiplex PCR in cooked ham. Food Microbiol. 2005, 22, 109–115. [Google Scholar]
- Gómez-Duarte, O.G.; Bai, J.; Newell, E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2009, 63, 1–9. [Google Scholar]
- Yaron, S.; Matthews, K.R. A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: Investigation of specific target genes. J. Appl. Microbiol. 2002, 92, 633–640. [Google Scholar]
- Morin, N.J.; Gong, Z.; Li, X.F. Reverse transcription-multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Vibrio cholerae O1, and Salmonella typhi. Clin. Chem. 2004, 50, 2037–2044. [Google Scholar]
- Liang, H.; Cordova, S.E.; Kieft, T.L.; Rogelj, S. A highly sensitive immuno-PCR assay for detecting group A Streptococcus. J. Immunol. Methods 2003, 279, 101–110. [Google Scholar]
- Zhang, W.; Bielaszewska, M.; Pulz, M.; Becker, K.; Friedrich, A.W.; Karch, H.; Kuczius, T. New immuno-PCR assay for detection of low concentrations of shiga toxin 2 and its variants. J. Clin. Microbiol. 2008, 46, 1292–1297. [Google Scholar]
- Cook, N. The use of NASBA for the detection of microbial pathogens in food and environmental samples. J. Microbiol. Methods 2003, 53, 165–174. [Google Scholar]
- Leonard, P.; Hearty, S.; Quinn, J.; O'Kennedy, R. A generic approach for the detection of whole Listeria monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens. Bioelectron. 2004, 19, 1331–1335. [Google Scholar]
- Leenaars, M.; Hendricksen, C.F.M. Critical steps in the production of polyclonal and monoclonal antibodies: Evaluation and recommendations. ILAR J. 2005, 46, 269–279. [Google Scholar]
- Arnold, J.N.; Wormald, M.R.; Sim, R.B.; Rudd, P.M.; Dwek, R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 2007, 25, 21–50. [Google Scholar]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature (London) 1975, 256, 495–497. [Google Scholar]
- Nelson, P.N.; Reynolds, G.M.; Waldron, E.E.; Ward, E.; Giannopoulos, K.; Murray, P.G. Monoclonal antibodies. Mol. Pathol. 2000, 53, 111–117. [Google Scholar]
- Bradbury, A.R.; Marks, J.D. Antibodies from phage antibody libraries. J. Immunol. Methods 2004, 290, 29–49. [Google Scholar]
- Hoogenboom, H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005, 23, 1105–1116. [Google Scholar]
- Finlay, W.J.; deVore, N.C.; Dobrovolskaia, E.N.; Gam, A.; Goodyear, C.S.; Slater, J.E. Exploiting the avian immunoglobulin system to simplify the generation of recombinant antibodies to allergenic proteins. Clin. Exp. Allergy 2005, 35, 1040–1048. [Google Scholar]
- Townsend, S.; Finlay, W.J.; Hearty, S.; O'Kennedy, R. Optimising recombinant antibody function in SPR immunosensing. The influence of antibody structural format and chip surface chemistry on assay sensitivity. Biosens. Bioelectron. 2006, 22, 268–274. [Google Scholar]
- Sakai, K.; Shimizu, Y.; Chiba, T.; Matsumoto-Takasaki, A.; Kusada, Y.; Zhang, W.; Nakata, M.; Kojima, N.; Toma, K.; Takayanagi, A.; Shimizu, N.; Fujita-Yamaguchi, Y. Isolation and characterisation of phage-displayed single chain antibodies recognizing nonreducing terminal mannose residues. 1. A new strategy for generation of anti-carbohydrate antibodies. Biochemistry 2007, 46, 253–262. [Google Scholar]
- Andris-Widhopf, J.; Steinberger, P.; Fuller, R.; Rader, C.; Barbas, C.F., III. Generation of antibody libraries: PCR amplification and assembly of light- and heavy-chain coding sequences. In Phage display: A laboratory manual, 1st; Barbas, C.F., Burton, D.R., Scott, J.K., Silverman, G.J., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 9.1–9.113. [Google Scholar]
- Hoogenboom, H.R.; Griffiths, A.D.; Johnson, K.S.; Chiswell, D.J.; Hudson, P.; Winter, G. Multi-subunit proteins on the surface of filamentous phage: Methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991, 19, 4133–4137. [Google Scholar]
- Maynard, J.; Georgiou, G. Antibody engineering. Annu. Rev. Biomed. Eng. 2000, 2, 339–376. [Google Scholar]
- Hudson, P.J.; Souriau, C. Engineered antibodies. Nat. Med. 2003, 9, 129–134. [Google Scholar]
- Leonard, P.; Säfsten, P.; Hearty, S.; McDonnell, B.; Finlay, W.; O'Kennedy, R. High throughput ranking of recombinant avian scFv antibody fragments from crude lysates using the Biacore A100. J. Immunol. Methods 2006, 323, 172–179. [Google Scholar]
- Lee, Y.C.; Leu, S.J.; Hu, C.J.; Shih, N.Y.; Huang, I.J.; Wu, H.H.; Hsieh, W.S.; Chiang, B.L.; Chiu, W.T.; Yang, Y.Y. Chicken single-chain variable fragments against the SARS-CoV spike protein. J. Virol. Methods 2007, 146, 104–111. [Google Scholar]
- Torensma, R.; Visser, M.J.C.; Aarsman, C.J.M.; Poppelier, M.J.J.G.; Fluit, A.C.; Verhoef, J. Monoclonal antibodies that react with live Listeria spp. Appl. Environ. Microbiol. 1993, 59, 2713–2716. [Google Scholar]
- Uyttendaele, M.; Van Hoorde, I.; Debevere, J. The use of immuno-magnetic separation (IMS) as a tool in a sample preparation method for direct detection of L. monocytogenes in cheese. Int. J. Food Microbiol. 2000, 54, 205–212. [Google Scholar]
- Jordan, D.; Vancov, T.; Chowdhury, A.; Andersen, L.M.; Jury, K.; Stevenson, A.E.; Morris, S.G. The relationship between concentration of a dual marker strain of Salmonella typhimurium in bovine faeces and its probability of detection by immunomagnetic separation and culture. J. Appl. Microbiol. 2004, 97, 1054–1062. [Google Scholar]
- Chapman, P.A.; Ashton, R. An evaluation of rapid methods for detecting Escherichia coli O157 on beef carcasses. Int. J. Food Microbiol. 2003, 87, 279–285. [Google Scholar]
- Kerr, P.; Finlay, D.; Thomson-Carter, F.; Ball, H.J. A comparison of a monoclonal antibody-based sandwich ELISA and immunomagnetic bead selective enrichment for the detection of Escherichia coli O157 from bovine faeces. J. Appl. Microbiol. 2001, 91, 933–936. [Google Scholar]
- Kerr, P.; Chart, H.; Finlay, D.; Pollock, D.A.; MacKie, D.P.; Ball, H.J. Development of a monoclonal sandwich ELISA for the detection of animal and human Escherichia coli O157 strains. J. Appl. Microbiol. 2001, 90, 543–549. [Google Scholar]
- Kim, S.H.; Park, M.K.; Kim, J.Y.; Chuong, P.D.; Lee, Y.S.; Yoon, B.S.; Hwang, K.K.; Lim, Y.K. Development of a sandwich ELISA for the detection of Listeria spp. using specific flagella antibodies. J. Vet. Sci. 2005, 6, 41–46. [Google Scholar]
- Tully, E.; Hearty, S.; Leonard, P.; O'Kennedy, R. The development of rapid fluorescence-based immunoassays, using quantum dot-labelled antibodies for the detection of Listeria monocytogenes cell surface proteins. Int. J. Biol. Macromol. 2006, 39, 127–134. [Google Scholar]
- Valdivieso-Garcia, A.; Riche, E.; Abubakar, O.; Waddell, T.E.; Brooks, B.W. A double antibody sandwich enzyme-linked immunosorbent assay for the detection of Salmonella using biotinylated monoclonal antibodies. J. Food Prot. 2001, 64, 1166–1171. [Google Scholar]
- MacBeath, G.; Schreiber, S.L. Printing proteins as microarrays for high-thoughput function determination. Science 2000, 289, 1760–1763. [Google Scholar]
- Gehring, A.G.; Albin, D.M.; Bhunia, A.K.; Reed, S.A.; Tu, S.I.; Uknalis, J. Antibody microarray detection of Escherichia coli O157:H7: Quantification, assay limitations, and capture efficiency. Anal. Chem. 2006, 78, 6601–6607. [Google Scholar]
- Cai, H.Y.; Lu, L.; Muckle, C.A.; Prescott, J.F.; Chen, S. Development of a novel protein microarray for serotyping Salmonella enterica strains. J. Clin. Microbiol. 2005, 43, 3427–3430. [Google Scholar]
- Anjum, M.F.; Tucker, J.D.; Sprigings, K.A.; Woodward, M.J.; Ehricht, R. Use of miniaturized protein arrays for Escherichia coli O serotyping. Clin. Vaccine Immunol. 2006, 13, 561–567. [Google Scholar]
- Gehring, A.G.; Albin, D.M.; Reed, S.A.; Tu, S.I.; Brewster, J.D. An antibody microarray, in multiwall plate format, for multiplex screening of foodborne pathogenic bacteria and biomolecules. Anal. Bioanal. Chem. 2008, 391, 497–506. [Google Scholar]
- Huelseweh, B.; Ehricht, R.; Marschall, H.J. A simple and rapid protein array based method for the simultaneous detection of biowarfare agents. Proteomics 2006, 6, 2972–2981. [Google Scholar]
- Sapsford, K.E.; Ngundi, M.M.; Moore, M.H.; Lassman, M.E.; Shriver-Lake, L.C.; Taitt, C.R.; Ligler, F.S. Rapid detection of foodborne contaminants using an array biosensor. Sens. Act. B 2006, 113, 599–607. [Google Scholar]
- Delehanty, J.B.; Ligler, F.S. A microarray immunoassay for simultaneous detection of proteins and bacteria. Anal. Chem. 2002, 74, 5681–5687. [Google Scholar]
- Palchetti, I.; Mascini, M. Electrochemical biosensors and their potential for food pathogen and toxin detection. Anal. Bioanal. Chem. 2008, 391, 455–471. [Google Scholar]
- Gehring, A.G.; Crawford, C.G.; Mazenko, R.S.; Van Houten, L.J.; Brewster, J.D. Enzyme-linked immunomagnetic electrochemical detection of Salmonella typhimurium. J. Immunol. Methods 1996, 195, 15–25. [Google Scholar]
- Ivnitski, D.; Wilkins, E.; Tien, H.T.; Ottova, A. Electrochemical biosensor based on supported planar lipid bilayers for fast detection of pathogenic bacteria. Electrochem. Comm. 2000, 2, 457–460. [Google Scholar]
- Lin, Y.H.; Chen, S.H.; Chuang, Y.C.; Lu, Y.C.; Shen, T.Y.; Chang, C.A.; Lin, C.S. Disposable amperometric immunosensing strips fabricated by Au nanoparticles-modified screen-printed carbon electrodes for the detection of foodborne pathogen Escherichia coli O157:H7. Biosens. Bioelectron. 2008, 23, 1832–1837. [Google Scholar]
- Crowley, E.L.; O'Sullivan, C.K.; Guilbault, G.G. Increasing the sensitivity of Listeria monocytogenes assays: Evaluation using ELISA and amperometric detection. Analyst 1999, 124, 295–299. [Google Scholar]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E.; Striker, S. Application of electrochemical biosensors for detection of food pathogenic bacteria. Electroanalysis 2000, 12, 317–325. [Google Scholar]
- Radke, S.M.; Alocilja, E.C. A high density microelectrode array biosensor for detection of E. coli O157:H7. Biosens. Bioelectron. 2005, 20, 1662–1667. [Google Scholar]
- Tully, E.; Higson, S.P.; O'Kennedy, R. The development of a ‘labeless’ immunosensor for the detection of Listeria monocytogenes cell surface protein, Internalin B. Biosens. Bioelectron. 2008, 23, 906–912. [Google Scholar]
- Wang, R.; Ruan, C.; Kanayeva, D.; Lassiter, K.; Li, Y. TiO2 nanowire bundle microelectrode-based impedance immunosensor for rapid and sensitive detection of Listeria monocytogenes. Nano Lett. 2008, 8, 2625–2631. [Google Scholar]
- Su, X.L.; Li, Y. A QCM immunosensor for Salmonella detection with simultaneous measurements of resonant frequency and motional resistance. Biosens. Bioelectron. 2005, 21, 840–848. [Google Scholar]
- Gehring, A.G.; Patterson, D.L.; Tu, S.I. Use of a light-addressable potentiometric sensor for the detection of Escherichia coli O157:H7. Anal. Biochem. 1998, 258, 293–298. [Google Scholar]
- Dill, K.; Stanker, L.H.; Young, C.R. Detection of Salmonella in poultry using a silicon chip-based biosensor. J. Biochem. Biophys. Methods 1999, 41, 61–67. [Google Scholar]
- Hoa, D.T.; Suresh Kumar, T.N.; Pnuekar, N.S.; Srinivasa, R.S.; Lal, R.; Contractor, A.Q. A biosensor based on conducting polymers. Anal. Chem. 1992, 64, 2645–2646. [Google Scholar]
- Muhammad-Tahir, Z.; Alocilja, E.C. A conductimetric biosensor for biosecurity. Biosens. Bioelectron. 2003, 18, 813–819. [Google Scholar]
- Muhammad-Tahir, Z.; Alocilja, E.C. A disposable biosensor for pathogen detection in fresh produce. Biosyst. Eng. 2004, 88, 145–151. [Google Scholar]
- Hnaiein, M.; Hassen, W.M.; Abdelghani, A.; Fournier-Wirth, C.; Coste, J.; Bessueille, F.; Leonard, D.; Jaffrezic-Renault, N. A conductometric immunosensor based on functionalised magnetite nanoparticles for E. coli detection. Electrochem. Commun. 2008, 10, 1152–1154. [Google Scholar]
- Ngeh-Ngwainbi, J.; Suleiman, A.A.; Guilbault, G.G. Piezoelectric crystal biosensors. Biosens. Bioelectron. 1990, 5, 13–26. [Google Scholar]
- Su, X.L.; Li, Y. A self-assembled monolayer-based piezoelectric immunosensor for rapid detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2004, 19, 563–574. [Google Scholar]
- Babacan, S.; Pivarnik, P.; Lecter, S.; Rand, A.G. Evaluation of antibody immobilization methods for piezoelectric biosensor application. Biosens. Bioelectron. 2000, 15, 615–621. [Google Scholar]
- Fung, Y.S.; Wong, Y.Y. Self-assembled monolayers as the coating in a quartz piezoelectric crystal immunosensor to detect Salmonella in aqueous solution. Anal. Chem. 2001, 73, 5302–5309. [Google Scholar]
- Kim, G.H.; Rand, A.G.; Letcher, S.V. Impedance characterization of a piezoelectric immunosensor part II: Salmonella typhimurium detection using magnetic enhancement. Biosens. Bioelectron. 2003, 18, 91–99. [Google Scholar]
- Pohanka, M.; Skládal, P.; Pavlis, O. Label-free piezoelectric immunosensor for rapid assay of Escherichia coli. J. Immunoassay Immunochem. 2008, 29, 70–79. [Google Scholar]
- Ramanathan, K.; Danielsson, B. Principles and applications of thermal biosensors. Biosens. Bioelectron. 2001, 16, 417–423. [Google Scholar]
- Ruan, C.; Zeng, K.; Varghese, O.K.; Grimes, C.A. Magnetoelastic immunosensors: Amplified mass immunosorbent assay for detection of Escherichia coli O157:H7. Anal. Chem. 2003, 75, 6494–6498. [Google Scholar]
- Mujika, M.; Arana, S.; Castaño, E.; Tijero, M.; Vilares, R.; Ruano-López, J.M.; Cruz, A.; Sainz, L.; Berganza, J. Magnetoresistive immunosensor for the detection of Escherichia coli O157:H7 including a microfluidic network. Biosens. Bioelectron. 2009, 24, 1253–1258. [Google Scholar]
- Kretschmann, E.; Raether, H. Radiative decay of non-radiative surface plasmons excited by light. Z. Naturforsch. 1968, 23, 2135–2136. [Google Scholar]
- Kretschmann, E. The determination of optical constants of metals by excitation of surface plasmon resonance. Z. Phys. 1971, 241, 313–324. [Google Scholar]
- Wei, D.; Oyarzabal, O.A.; Huang, T.S.; Balasubramanian, S.; Sista, S.; Simonian, A.L. Development of a surface plasmon resonance biosensor for the identification of Campylobacter jejuni. J. Microbiol. Methods 2007, 69, 78–85. [Google Scholar]
- Barlen, B.; Mazumdar, S.D.; Lezrich, O.; Kämpfer, P.; Keugsen, M. Detection of Salmonella by surface plasmon resonance. Sensors 2007, 7, 1427–1446. [Google Scholar]
- Mazumdar, S.D.; Hartmann, M.; Kämpfer, P.; Keugsen, M. Rapid method for detection of Salmonella in milk by surface plasmon resonance. Biosens. Bioelectron. 2007, 22, 2040–2046. [Google Scholar]
- Oh, B.K.; Lee, W.; Chun, B.S.; Bae, Y.M.; Lee, W.H.; Choi, J.W. The fabrication of protein chip based on surface plasmon resonance for detection of pathogens. Biosens. Bioelectron. 2005, 20, 1847–1850. [Google Scholar]
- Koubová, V.; Brynda, E.; Karasová, L.; Škvor, J.; Homola, J.; Dostálek, J.; Tobiška, P.; Rošicky, J. Detection of foodborne pathogens using surface plasmon resonance biosensors. Sens. Act. B 2001, 74, 100–105. [Google Scholar]
- Taylor, A.D.; Ladd, J.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar]
- Rijal, K.; Leung, A.; Mohana Shankar, P.; Mutharasan, R. Detection of pathogenic Escherichia coli O157:H7 at 70 cells/mL using antibody-immobilized biconical tapered fiber sensors. Biosens. Bioelectron. 2005, 21, 871–880. [Google Scholar]
- Donaldson, K.A.; Kramer, M.F.; Lim, D.V. A rapid detection method for Vaccinia virus, the surrogate for smallpox virus. Biosens. Bioelectron. 2004, 20, 322–327. [Google Scholar]
- Lim, D.V. Detection of microorganisms and toxins with evanescent wave fiber-optic biosensors. Proc. IEEE 2003, 91, 902–907. [Google Scholar]
- Anderson, G.P.; Nerurkar, N.L. Improved fluoroimmunoassays using the dye Alexa Fluor 647 with the RAPTOR, a fiber optic biosensor. J. Immunol. Methods 2002, 271, 17–24. [Google Scholar]
- Kramer, M.F.; Lim, D.V. A rapid and automated fiber optic-based biosensor assay for the detection of Salmonella in spent irrigation water used in the sprouting of sprout seeds. J. Food Prot. 2004, 67, 46–52. [Google Scholar]
- Nanduri, V.; Kim, G.; Morgan, M.T.; Ess, D.; Hahm, B.K.; Kothapalli, A.; Valadez, A.; Geng, T.; Bhunia, A.K. Antibody immobilization on waveguides using a flow-through system shows improved Listeria monocytogenes detection in an automated fiber optic biosensor: RAPTOR™. Sensors 2006, 6, 808–822. [Google Scholar]
- Leskinen, S.D.; Lim, D.V. Rapid ultrafiltration concentration and biosensor detection of Enterococci from large volumes of Florida recreational water. Appl. Env. Microbiol. 2008, 74, 4792–4798. [Google Scholar]
- Fratamico, P.M.; Strobaugh, T.P.; Medina, M.B.; Gehring, A.G. Detection of Escherichia coli O157:H7 using a surface plasmon resonance biosensor. Biotechnol. Techn. 1998, 12, 571–576. [Google Scholar]
- Bokken, G.C.; Corbee, R.J.; van Knapen, F.; Bergwerff, A.A. Immunochemical detection of Salmonella group B, D and E using an optical surface plasmon resonance biosensor. FEMS Microbiol. Lett. 2003, 222, 75–82. [Google Scholar]
- Haines, J.; Patel, P.D. Detection of food borne pathogens using BIA. BIA J. 1995, 2, 31. [Google Scholar]
- Harteveld, J.L.N.; Nieuwenhuizen, M.S.; Wils, E.R.J. Detection of Staphylococcal enterotoxin B employing a piezoelectric crystal immunosensor. Biosens. Bioelectron. 1997, 12, 661–667. [Google Scholar]
- Slavík, R.; Homola, J.; Brynda, E. A miniature fiber optic surface plasmon resonance sensor for fast detection of Staphylococcal enterotoxin B. Biosens. Bioelectron. 2002, 17, 591–595. [Google Scholar]
- Moreno-Bondi, M.C.; Rowe-Taitt, C.; Shriver-Lake, L.C.; Ligler, F.S. Multiplexed measurement of serum antibodies using an array biosensor. Biosens. Bioelectron. 2006, 21, 1880–1886. [Google Scholar]
- Subramanian, A.; Irudayaraj, J.; Ryan, T. Mono and dithiol surfaces on surface plasmon resonance biosensors for detection of Staphylococcus aureus. Sens. Act. B 2006, 114, 192–198. [Google Scholar]
- Tims, T.B.; Lim, D.V. Rapid detection of Bacillus anthracis spores directly from powders from an evanescent wave fiber-optic biosensor. J. Microbiol. Methods 2004, 59, 127–130. [Google Scholar]
- Campbell, G.A.; Mutharasan, R. Method of measuring Bacillus anthracis spores in the presence of copious amounts of Bacillus thuringiensis and Bacillus cereus. Anal. Chem. 2007, 79, 1145–1152. [Google Scholar]
- Song, J.M.; Culha, M.; Kasili, P.M.; Griffin, G.D.; Vo-Dinh, T. A compact CMOS biochip immunosensor towards the detection of a single bacteria. Biosens. Bioelectron. 2005, 20, 2203–2209. [Google Scholar]
- Uithoven, K.A.; Schmidt, J.C.; Ballman, M.E. Rapid identification of biological warfare agents using an instrument employing a light addressable potentiometric sensor and a flow-through immunofiltration-enzyme assay system. Biosens. Bioelectron. 2000, 14, 761–770. [Google Scholar]
- Meyer, M.H.F.; Krause, H.J.; Hartmann, M.; Miethe, P.; Oster, J.; Keusgen, M. Francisella tularensis detection using magnetic labels and a magnetic biosensor based on frequency mixing. J. Magn. Mag. Mat. 2007, 311, 259–263. [Google Scholar]
- He, F.; Zhang, L. Rapid diagnosis of M. tuberculosis using a piezoelectric immunosensor. Anal. Sci. 2002, 18, 397–401. [Google Scholar]
- Díaz-González, M.; González-García, M.B.; Costa-García, A. Immunosensor for Mycobacterium tuberculosis on screen-printed carbon electrodes. Biosens. Bioelectron. 2005, 20, 2035–2043. [Google Scholar]
- García-Ojeda, P.A.; Hardy, S.; Kozlowski, S.; Stein, K.E.; Feavers, I.M. Surface plasmon resonance analysis of antipolysaccharide antibody specificity: Responses to meningococcal group C conjugate vaccines and bacteria. Infect. Immun. 2004, 72, 3451–3460. [Google Scholar]
- Rao, V.K.; Sharma, M.K.; Goel, A.K.; Singh, L.; Sekhar, K. Amperometric immunosensor for the detection of Vibrio cholerae O1 using disposable screen-printed electrodes. Anal. Sci. 2006, 22, 1207–1211. [Google Scholar]
- Jyoung, J.Y.; Hong, S.; Lee, W.; Choi, J.W. Immunosensor for the detection of Vibrio cholerae O1 using surface plasmon resonance. Biosens. Bioelectron. 2006, 21, 2315–2319. [Google Scholar]
- Meyer, M.H.F.; Stehr, M.; Bhuju, S.; Krause, H.J.; Hartmann, M.; Miethe, P.; Singh, M.; Keusgen, M. Magnetic biosensor for the detection of Yersinia pestis. J. Microbiol. Methods 2007, 68, 218–224. [Google Scholar]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar]
- Skottrup, P.; Nicolaisen, M.; Justesen, A.F. Rapid determination of Phytophthora infestans sporangia using a surface plasmon resonance immunosensor. J. Immunol. Methods 2007, 68, 507–515. [Google Scholar]
- Skottrup, P.; Hearty, S.; Frøkiaer, H.; Leonard, P.; Hejgaard, J.; O'Kennedy, R.; Nicolaisen, M.; Justesen, A.F. Detection of fungal spores using generic surface plasmon resonance immunoassay. Biosens. Bioelecton. 2007, 22, 2724–2729. [Google Scholar]
- Muramatsu, H.; Kajiwara, K.; Tamiya, E.; Karube, I. Piezoelectric immuno sensor for the detection of Candida albicans microbes. Anal. Chim. Acta 1986, 188, 257–261. [Google Scholar]
- Medyantseva, E.P.; Khaldeeva, E.V.; Glushko, N.I.; Budnikov, H.C. Amperometric enzyme immunosensor for the determination of the antigen of the pathogenic fungi Trichophyton rubrum. Anal. Chim. Acta 2000, 411, 13–18. [Google Scholar]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar]
- Daly, S.J.; Keating, G.J.; Dillon, P.P.; Manning, B.M.; O'Kennedy, R.; Lee, H.A.; Morgan, M.R.A. Development of surface plasmon resonance-based immunoassay for aflatoxin B1. J. Agric. Food Chem. 2000, 48, 5097–5104. [Google Scholar]
- Daly, S.J.; Dillon, P.P.; Manning, B.M.; Dunne, L.; Killard, A.; O'Kennedy, R. Production and characterisation of murine single chain Fv antibodies to aflatoxin B-1 derived from a pre-immunised antibody phage display library system. Food Agric. Immunol. 2002, 14, 255–274. [Google Scholar]
- Dunne, L.; Daly, S.; Baxter, A.; Haughey, S.; O'Kennedy, R. Surface plasmon resonance-based immunoassay for the detection of aflatoxin B-1 using single-chain antibody fragments. Spectroscopy Lett. 2005, 38, 229–245. [Google Scholar]
- Adányi, N.; Levkovets, I.A.; Rodriguez-Gil, S.; Ronald, A.; Váradi, M.; Szendro, I. Development of immunosensor based on OWLS technique for determining aflatoxin B1 and ochratoxin A. Biosens. Bioelectron. 2007, 22, 797–802. [Google Scholar]
- Sournia, A.; Chrdtiennot-Dinet, M.J.; Ricard, M. Marine phytoplankton: How many species in the world ocean? J. Plankton Res. 1991, 13, 1093–1099. [Google Scholar]
- Van Egmond, H.P.; Speijers, G.J.A.; Van den Top, H.J. Current situation on worldwide regulations for marine phycotoxins. J. Natural Toxins 1992, 1, 67–85. [Google Scholar]
- Koenig, B.; Graetzel, M. A novel immunosensor for herpes viruses. Anal. Chem. 1994, 66, 341–344. [Google Scholar]
- Rickert, J.; Göpel, W.; Beck, W.; Jung, G.; Heiduschka, P. A ‘mixed’ self-assembled monolayer for an impedimetric immunosensor. Biosens. Bioelectron. 1996, 11, 757–768. [Google Scholar]
- Uttenthaler, E.; Kösslinger, R.; Drost, S. Characterisation of immobilization methods for African swine fever virus protein and antibodies with a piezoelectric immunosensor. Biosens. Bioelectron. 1998, 13, 1279–1286. [Google Scholar]
- Ditcham, W.G.; Al-Obaidi, A.H.; McStay, D.; Mottram, T.T.; Brownlie, J.; Thompson, I. An immunosensor with potential for the detection of viral antigens in body fluids, based on surface second harmonic generation. Biosens. Bioelectron. 2001, 16, 221–224. [Google Scholar]
- Eun, A.J.; Huang, L.; Chew, F.T.; Li, S.F.; Wong, S.M. Detection of two orchid viruses using quartz crystal microbalance (QCM) immunosensors. J. Virol. Methods 2002, 99, 71–79. [Google Scholar]
- Zuo, B.; Li, S.; Guo, Z.; Zhang, J.; Chen, C. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal. Chem. 2004, 76, 3536–3540. [Google Scholar]
- Minunni, M.; Tombelli, S.; Gullotto, A.; Luzi, E.; Mascini, M. Development of biosensors with aptamers as bio-recognition element: The case of HIV-1 Tat protein. Biosens. Bioelectron. 2004, 20, 1149–1156. [Google Scholar]
- Konry, T.; Novoa, A.; Shemer-Avni, Y.; Hanuka, N.; Cosnier, S.; Lepellec, A.; Marks, R.S. Optical fiber immunosensor based on a poly(pyrrole-benzophenone) film for the detection of antibodies to viral antigen. Anal. Chem. 2005, 1771–1779. [Google Scholar]
- Torrance, L.; Ziegler, A.; Pittman, H.; Paterson, M.; Toth, R.; Eggleston, I. Oriented immobilisation of engineered single-chain antibodies to develop biosensors for virus detection. J. Virol. Methods 2006, 134, 164–170. [Google Scholar]
- Yu, J.S.; Liao, H.X.; Gerdon, A.E.; Huffman, B.; Scearce, R.M.; McAdams, M.; Alam, S.M.; Popernack, P.M.; Sullivan, N.J.; Wright, D.; Cliffel, D.E.; Nabel, G.J.; Haynes, B.F. Detection of Ebola virus envelope using monoclonal and polyclonal antibodies in ELISA, surface plasmon resonance and a quartz crystal microbalance immunosensor. J. Virol. Methods 2006, 137, 219–228. [Google Scholar]
- Huang, J.G.; Lee, C.L.; Lin, H.M.; Chuang, T.L.; Wang, W.S.; Juang, R.H.; Wang, C.H.; Lee, C.K.; Lin, S.M.; Lin, C.W. A miniaturized germanium-doped silicon dioxide-based surface plasmon resonance waveguide sensor for immunoassay detection. Biosens. Bioelectron. 2006, 22, 519–525. [Google Scholar]
- Sobarzo, A.; Paweska, J.T.; Herrmann, S.; Amir, T.; Marks, R.S.; Lobel, L. Optical fiber immunosensor for the detection of IgG antibody to Rift Valley fever virus in humans. J. Virol. Methods 2007, 146, 327–334. [Google Scholar]
- Ionescu, R.E.; Cosnier, S.; Herrmann, S.; Marks, R.S. Amperometric immunosensor for the detection of anti-West Nile virus IgG. Anal. Chem. 2007, 79, 8662–8668. [Google Scholar]
- Lehane, L. Paralytic shellfish poisoning: A review; National Office of Animal and Plant Health, Agriculture, Fisheries and Forestry: Australia, 2000. [Google Scholar]
- Pan, Y.; Parsons, M.L.; Busman, M.; Moeller, P.D.R.; Dortch, Q.; Powell, C.L.; Douchette, G.J. Pseudo-nitzschia spp. of Pseudodelicatissima- a confirmed producer of domoic acid from the northern gulf of Mexico. Marine Ecol. Prog. Ser. 2001, 220, 83–92. [Google Scholar]
- Bowden, B.F. Yessotoxins-polycyclic ethers from dinoflagellates: Relationships to diarrhetic shellfish toxins. Toxin Rev. 2006, 25, 137–157. [Google Scholar]
- Satake, M.; MacKenzie, L.; Yasumoto, T. Identification of Protoceratium reticulatum as a biogenetic origin of yessotoxin. Nat. Toxins 1997, 5, 164–167. [Google Scholar]
- Falconer, I.R.; Humpage, A.R. Health risk assessment of cyanobacterial (blue-green algal) toxins in drinking water. Int. J. Environ. Public Health 2005, 2, 43–50. [Google Scholar]
- Hirama, M. Total synthesis of ciguatoxin CTX3C: A venture into the problems of ciguatera seafood poisoning. The Chem. Rec. 2005, 5, 240–250. [Google Scholar]
- Carter, R.M.; Poli, M.A.; Pesavento, M.; Sibley, D.E.T.; Lubrano, G.J.; Guilbault, G.G. Immunoelectrochemical biosensors for detection of saxitoxin and brevetoxin. Immunomethods 1993, 3, 128–133. [Google Scholar]
- Kreuzer, M.P.; Pravda, M.; O'Sullivan, C.K.; Guilbault, G.G. Novel electrochemical immunosensors for seafood toxin analysis. Toxicon 2002, 40, 1267–1274. [Google Scholar]
- Kania, M.; Kreuzer, M.; Moore, E.; Pravda, M.; Hock, B.; Guilbault, G. Development of polyclonal antibodies against domoic acid for their use in electrochemical biosensors. Anal. Lett. 2003, 36, 1851–1863. [Google Scholar]
- Lotierzo, M.; Henry, O.Y.; Piletsky, S.; Tothill, I.; Cullen, D.; Kania, M.; Hock, B.; Turner, A.P. Surface plasmon resonance sensor for domoic acid based on grafted imprinted polymer. Biosens. Bioelectron. 2004, 20, 145–152. [Google Scholar]
- Yu, Q.; Chen, S.; Taylor, A.D.; Homola, J.; Hock, B.; Jiang, S. Detection of low-molecular-weight domoic acid using surface plasmon resonance sensor. Sens. Act. B 2005, 107, 193–201. [Google Scholar]
- Stevens, R.C.; Soelberg, S.D.; Eberhart, B.T.L.; Spencer, S.; Wekell, J.C.; Chinowsky, T.M.; Trainer, V.L.; Furlong, C.E. Detection of the toxin domoic acid from clam extracts using a portable surface plasmon resonance biosensor. Harm. Algae 2007, 6, 166–174. [Google Scholar]
- Long, F.; He, M.; Shi, H.C.; Zhu, A.N. Development of evanescent wave all-fiber immunosensor for environmental water analysis. Biosens. Bioelectron. 2008, 23, 952–958. [Google Scholar]
- Loyprasert, S.; Thavarungkul, P.; Asawatreratanakul, P.; Wongkittisuksa, B.; Limsakul, C.; Kanatharana, P. Label-free capacitive immunosensor for microcystin-LR using self-assembled thiourea monolayer incorporated with Ag nanoparticles on gold electrode. Biosens. Bioelectron. 2008, 24, 78–86. [Google Scholar]
- Long, F.; He, M.; Zhu, A.N.; Shi, H.C. Portable optical immunosensor for highly sensitive detection of microcystin-LR in water samples. Biosens. Bioelectron. 2009, 24, 2346–2351. [Google Scholar]
- Marquette, C.A.; Coulet, P.R.; Blum, L.J. Semi-automated membrane based on chemiluminescent immunosensor for flow injection analysis of okadaic acid in mussels. Anal. Chim. Acta 1999, 398, 173–182. [Google Scholar]
- Tang, A.X.J.; Pravda, M.; Guilbault, G.G.; Piletsky, S.; Turner, A.P.F. Immunosensor for okadaic acid using quartz crystal microbalance. Anal. Chim. Acta 2002, 471, 33–40. [Google Scholar]
- Tang, A.; Kreuzer, M.; Lehane, M.; Pravda, M.; Guilbault, G.G. Immunosensor for the determination of okadaic acid based on screen-printed electrode. Int. J. Environ. Anal. Chem. 2003, 83, 663–670. [Google Scholar]
- Campás, M.; de la Igleisa, P.; Le Berre, M.; Kane, M.; Diogéne, J.; Marty, J.L. Enzymatic recycling-based amperometric immunosensor for the ultrasensitive detection of okadaic acid in shellfish. Biosens. Bioelectron. 2008, 24, 716–722. [Google Scholar]
- Satake, M.; Ofuki, K.; Naoki, H.; James, K.J.; Furey, A.; McMahon, T.; Silke, J.; Yasumoto, T. Azaspiracid, a new marine toxin having unique spiro ring assemblies, isolated from Irish mussels, Mytilus edulis. J. Am. Chem. Soc. 1998, 120, 9967–9968. [Google Scholar]
- Twiner, M.J.; Rehmann, N.; Hess, P.; Douchette, G.J. Azaspiracid shellfish poisoning: A review on the chemistry, ecology, and toxicology with an emphasis on human health impacts. Mar. Drugs 2008, 6, 39–72. [Google Scholar]
- Hess, P.; Nguyen, L.; Aasen, J.; Keogh, M.; Kilcoyne, J.; McCarron, P.; Aune, T. Tissue distribution, effects of cooking and parameters affecting the extraction of azaspiracids from mussels, Mytilus edulis, prior to analysis by liquid chromatography coupled with mass spectrometry. Toxicon 2005, 46, 62–71. [Google Scholar]
- Forsyth, C.J.; Xu, J.; Nguyen, S.T.; Samdal, I.A.; Briggs, L.R.; Rundberget, T.; Sandvik, M.; Miles, C.O. Antibodies with broad specificity to azaspiracids by use of synthetic haptens. J. Am. Chem. Soc. 2006, 128, 15114–15116. [Google Scholar]
- Campbell, G.A.; Mutharasan, R. Near real-time detection of Cryptosporidium parvum oocyst by IgM-functionalized piezoelectric-excited millimeter-sized cantilever biosensor. Biosens. Bioelectron. 2008, 23, 1039–1045. [Google Scholar]
- Kang, C.D.; Cao, C.; Lee, J.; Choi, I.S.; Kim, B.W.; Sim, S.J. Surface plasmon resonance-based inhibition assay for real-time detection of Cryptosporidium parvum oocyst. Water Res. 2008, 42, 1693–1699. [Google Scholar]
- Zhou, Y.M.; Liu, G.D.; Wu, Z.Y.; Shen, G.L.; Yu, R.Q. An amperometric immunosensor based on a conducting immunocomposite electrode for the determination of Schistosoma japonicum antigen. Anal. Sci. 2002, 18, 155–159. [Google Scholar]
- Zhou, Y.M.; Wu, Z.Y.; Shen, G.L.; Yu, R.Q. An amperometric immunosensor based on nafion-modified electrode for the detection of Schistosoma japonicum antibody. Sens. Act. B 2003, 89, 292–298. [Google Scholar]
- Zhong, T.S.; Liu, G. Silica sol-gel amperometric immunosensor for Schistosoma japonicum antibody assay. Anal. Sci. 2004, 20, 537–541. [Google Scholar]
- Wu, Z.; Wu, J.; Wang, S.; Shen, G.; Yu, R. An amplified mass piezoelectric immunosensor for Schistosoma japonicum. Biosens. Bioelectron. 2006, 22, 207–212. [Google Scholar]
- Nagel, T.; Gajovic-Eichelmann, N.; Tobisch, S.; Schulte-Spechtel, U.; Bier, F.F. Serodiagnosis of Lyme borreliosis infection using surface plasmon resonance. Clin. Chim. Acta 2008, 394, 110–113. [Google Scholar]
- Hahm, B.K.; Bhunia, A.K. Effect of environmental stresses on antibody-based detection of Escherichia coli O157:H7, Salmonella enterica serotype Enteritidis and Listeria monocytogenes. J. Appl. Microbiol. 2006, 100, 1017–1027. [Google Scholar]
- Needham, R.; Williams, J.; Beales, N.; Voysey, P.L.; Magan, N. Early detection and differentiation of spoilage of bakery products. Sens. Act. B 2005, 106, 20–23. [Google Scholar]
- Alocilja, E.C.; Ritchie, N.L.; Grooms, D. L. Protocol development using an electronic nose for differentiating E. coli strains. IEEE Sens. J. 2003, 3, 801–805. [Google Scholar]
- Balasubramanian, S.; Panigrahi, S.; Logue, C.M.; Marchello, M.; Sherwood, J.S. Identification of Salmonella-inoculated beef using a portable electronic nose system. J. Rapid Methods Auto. Microbiol. 2005, 13, 71–95. [Google Scholar]
- Scampicchio, M.; Ballabio, D.; Arecchi, A.; Cosio, S.M.; Mannino, S. Amperometric electronic tongue for food analysis. Microchim. Acta. 2008, 163, 11–21. [Google Scholar]
- Lan, Y.B.; Wang, S.Z.; Yin, Y.G.; Hoffmann, C.W.; Zheng, X.Z. Using a surface plasmon resonance biosensor for rapid detection of Salmonella typhimurium in chicken carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar]
- Ngundi, M.M.; Kulagina, N.V.; Anderson, G.P.; Taitt, C.R. Nonantibody-based recognition: Alternative molecules for detection of pathogens. Exp. Rev. Proteomics 2006, 3, 511–524. [Google Scholar]
- Mascini, M.; Macagnano, A.; Monti, D.; Del Carlo, M.; Paolesse, R.; Chen, B.; Warner, P.; D'Amico, A.; Di Natale, C.; Compagnone, D. Piezoelectric sensors for dioxins: A biomimetic approach. Biosens. Bioelectron. 2004, 20, 1203–1210. [Google Scholar]
- Kulagina, N.V.; Shaffer, K.M.; Anderson, G.P.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptide-based array for Escherichia coli and Salmonella screening. Anal. Chim. Acta 2006, 575, 9–15. [Google Scholar]
- Pan, Q.; Zhang, X.L.; Wu, H.Y. Aptamers that preferentially bind type IVB pili and inhibit human monocytic-cell invasion by Salmonella enterica serovar typhi. Antimicrob. Agents Chemother. 2006, 49, 4052–4060. [Google Scholar]
- Mazarei, M.; Teplova, I.; Hajimorad, M.R.; Stewart, C.N., Jr. Pathogen phytosensing: Plants to report plant pathogens. Sensors 2008, 8, 2628–2641. [Google Scholar]







| Pathogen | Pathogenic trait |
|---|---|
| Bacterial Pathogens | |
| Bacillus anthracis | Human pathogen; causative agent of anthrax; toxin producer |
| Bacillus subtilis | Putative human pathogen: causative agent of food poisoning |
| Brucella abortus | Human and animal pathogen; causative agent of brucellosis |
| Campylobacter spp. and C. jejuni | Human pathogen; causative agent of campylobacteriosis |
| Clostridium botulinum | Human pathogen; producer of neurotoxins and causative agent of botulism |
| Escherichia coli O157:H7 | Human pathogen; causative agent of foodborne illness and producer of toxins, such as verocytoxin or ‘shiga-like’ toxin |
| Francisella tularensis | Animal pathogen; putative biohazard |
| Legionella pneumophila | Human pathogen; causative agent of Legionnaires disease (legionellosis) |
| Listeria monocytogenes | Human pathogen; causative agent of listeriosis |
| Mycobacterium tuberculosis | Human pathogen; causative agent of tuberculosis |
| Neisseria meningitidis | Human pathogen; causative agent of bacterial meningitis |
| Salmonella typhimurium | Human pathogen; causative agent of salmonellosis |
| Staphylococcus aureus | Human pathogen; causative agent of hospital-acquired infection, toxin producer |
| Yersinia enterocolitica | Human pathogen; causative agent of yersiniosis |
| Yersinia pestis | Human pathogen; potential causative agent of the black plague |
| Fungal pathogens | |
| Candida albicans | Human pathogen; causative agent of vaginal thrush |
| Puccinia striiformis | Plant pathogen; causative agent of stripe rust |
| Phytophthora infestans | Plant pathogen; causative agent of potato blight |
| Trichophyton rubrum | Human pathogen; causative agent of athlete's foot and ringworm |
| Viral pathogens | |
| African swine fever virus | Animal pathogen; causative agent of African swine fever |
| Bovine diarrhoea virus | Animal pathogen; causative agent of mucosal erosion and bovine diarrhoea |
| Cowpea mosaic virus | Plant pathogen; causes mosaic pattern, vein yellowing and leaf malformation |
| Ebola virus | Human pathogen; causative agent of severe haemorrhagic fever disease |
| Foot and mouth virus | Animal pathogen; causative agent of acute degenerative disease in cattle |
| Hepatitis C virus | Human pathogen; causative agent of blood-borne infectious disease |
| Human immunodeficiency virus | Human pathogen; causative agent of acquired immunodeficiency syndrome (AIDS) |
| Rift valley fever virus | Animal pathogen; causative agent of Rift valley fever |
| SARS-associated coronavirus | Human and animal pathogen; causative agent of severe acute respiratory syndrome |
| Tobacco mosaic virus | Plant virus; causes mottling and discolouration of leaves |
| West Nile virus | Human and animal virus; causative agent of West Nile fever and encephalitis |
| Strain and morphology | Selective media | Clinical signs of infection | Estimated annual cases * | Infectious doses (CFU) * |
|---|---|---|---|---|
| E. coli O157:H7 Gram negative rod | Cefixime rhamnose sorbitol MacConkey agar [4] SEL media [5]. | Diarrhoea (bloody) Renal failure Haemolytic uraemic syndrome (rare) | 173,107 | 1 × 101 - 1 × 102 |
| Salmonella spp. Gram negative rod | Bismuth sulphide agar [4] SEL media [5] | Cramps Diarrhoea Vomiting | 1,342,532 | 1 × 104 - 1 × 107 |
| L. monocytogenes Gram negative rod | Listeria enrichment broth [4,6] Fraser broth [4] SEL media [5] | Vomiting Abdominal cramps Fever | 2,493 | 400 - 1 × 103 |
| Technique | Pathogen application | Ref. |
|---|---|---|
| Real-time PCR | Mycobacterium avium subsp. Paratuberculosis | [15] |
| E. coli O157:H7 | [16] | |
| S. aureus | [17] | |
| L. monocytogenes | [8,18] | |
| S. enterica serovar typhimurium | [19] | |
| Multiplex PCR | E. coli O157:H7; Salmonella spp.; Shigella spp. | [20] |
| L. monocytogenes and Salmonella spp. | [21] | |
| Campylobacter spp., Salmonella spp., E. coli, Shigella spp., Vibrio cholerae, Y. enterocolitica | [22] | |
| Reverse transcriptase PCR | E. coli O157:H7 | [23] |
| E. coli O157:H7, V. cholerae, S. typhi | [24] | |
| Immuno PCR | Streptococcus pyogenes | [25] |
| E. coli shiga-toxin 2 | [26] | |
| NASBA | L. monocytogenes | [12,13] |
| Campylobacter spp., L. monocytogenes, S. enterica serovar Enteritidis | [27] |
| Bacterial strain | Biosensor format | Assay format | Antibodies | Sensitivity | Ref. |
|---|---|---|---|---|---|
| B. anthracis | Optical | Sandwich | Biotinylated rabbit anti-B. anthracis polyclonal [C]; rabbit anti-B. anthracis polyclonal CY5 [D] | 3.2 × 105 spores/mg powder | [108] |
| Piezoelectric | Capture | Rabbit polyclonal anti-B. anthracis [C] | 333 spores/mL | [109] | |
| B. globigii | Optical | Sandwich | Goat anti-B. globigii [C]; rabbit anti-B. globigii [S]; goat anti-rabbit-AP [D] | 1 spore | [110] |
| B. subtilis | Potentiometric | Sandwich | Biotinylated polyclonal anti-B. subtilis antibody [C]; fluorescein-labelled polyclonal anti-B. subtilis antibody [S]; anti-fluorescein urease-conjugated antibody [D] | 3 × 103 spores/mL | [111] |
| F. tularensis | Magnetic | Sandwich | Monoclonal anti-F. tularensis [C]; biotinylated monoclonal anti-F. tularensis on streptavidin-coated magnetic beads [D] | 1 × 104 – 1 × 106 CFU/mL | [112] |
| M. tuberculosis | Piezoelectric | Capture | Rabbit anti-M. tuberculosis [C] | 1 × 105 cells/mL | [113] |
| Voltammetric | Sandwich | Biotinylated rabbit anti-M. tuberculosis [C]; murine monoclonal anti-M. tuberculosis [S]; rabbit anti-mouse-AP [D] | 1.0 ng/mL | [114] | |
| N. meningitidis | Optical | Direct | Murine anti-group C polysaccharide [C] | - | [115] |
| V. cholerae | Amperometric | Sandwich | Rabbit polyclonal anti-V. cholerae [C]; mouse anti-V. cholerae [S]; anti-mouse AP [D] | 1 × 105 cells/mL | [116] |
| Optical | Capture | Monoclonal anti-V. cholerae O1 [C] | 1 × 105 – 1 × 109 cells/mL | [117] | |
| Y. pestis | Magnetic | Sandwich | Monoclonal anti-F1 antigen [C]; biotinylated monoclonal anti-F1 on streptavidin-coated magnetic beads [D] | 2.5 ng/mL antigen | [118] |
| Virus | Biosensor platform | Assay format | Antibodies | Ref. |
|---|---|---|---|---|
| Herpes simplex virus (HSV) 1 and 2, Varicella-Zoster virus (VSV), Cytomegalovirus (CMV) and Epstein-Barr virus (EBV) | Piezoelectric | Capture | Mouse monoclonal antibodies to herpes simplex virus 1 and 2, cytomegalovirus, Epstein-Barr virus and Varicella Zoster virus [C] | [131] |
| Foot and mouth virus (FMV) | Impedimetric | Indirect | Murine monoclonal [P] | [132] |
| African swine fever virus (ASF) | Piezoelectric | Capture | Murine monoclonal [C] | [133] |
| Bovine diarrhoeal virus (BVD) | Optoelectronic | Capture | Anti-BVD monoclonal | [134] |
| Cymbidium mosaic potexvirus (CymMV) and Odontoglossum ringspot tobamovirus (ORSV) | Piezoelectric | Capture | Rabbit polyclonal | [135] |
| SARS-associated coronavirus (SARS-CoV) | Piezoelectric | Capture | Horse polyclonal anti-SARS-CoV [C] | [136] |
| Human immunodeficiency virus (HIV-1) | Piezoelectric | Capture | Murine anti-trans activator of transcription (TAT) HIV [C] | [137] |
| Hepatitis C virus (HCV) | Optical | Indirect | Polyclonal IgG antibodies [P]; Polyclonal goat anti-human IgG-HRP [D] | [138] |
| Cowpea mosaic virus (CPMV) | Optical | Capture | Anti-CPMV recombinant antibody (scFv) fused to the constant light chain (CL) domain containing a C-terminal cysteine residue [C] | [139] |
| Ebola virus (EBOV) | Optical | Capture | Mouse monoclonal anti-EBOV [C] | [140] |
| QCM | Capture | Rabbit polyclonal antibody [C] or Mouse monoclonal antibody [C] | ||
| Avian leucosis virus (ALV) | Optical | Capture | Monoclonal anti-ALV-J | [141] |
| Rift valley fever virus (RVF) | Fibre optic immunosensor | Sandwich | Mouse polyclonal anti-RVF [C]; Polyclonal IgG antibodies [S]; Goat anti-human IgG – HRP [D] | [142] |
| West Nile virus (WNV) | Amperometric | Indirect | Polyclonal IgG antibodies [P]; Goat anti-human IgG-HRP [D] | [143] |
| Toxin | Biosensor Format | Assay Format | Antibodies | LOD | Ref |
|---|---|---|---|---|---|
| Brevetoxin | Amperometric | Indirect | Goat-anti brevetoxin [P] | 15 μg/L | [150] |
| Domoic acid (DA) | Amperometric | Indirect | Sheep polyclonal [P]; anti-sheep IgG-AP [D] | 2 μg/L | [151] |
| Amperometric | Indirect | Rabbit polyclonal [P] | 0.1 μg/L | [152] | |
| Optical | Indirect | Monoclonal anti-DA [P] | 1.8 μg/L | [153] | |
| Optical | Indirect | Monoclonal anti-DA [P] | 0.1 μg/L | [154] | |
| Optical | Indirect | Rabbit polyclonal anti-DA [P] | 3 μg/L | [155] | |
| Microcystin-LR (MC) | Optical | Direct | Monoclonal anti-MC-LR-Cy5 [P] | 0.03 μg/L | [156] |
| Capacitance | Capture | Monoclonal anti-MC-LR [C] | 7 pg/L | [157] | |
| Optical | Direct | Monoclonal anti-MC-LR-Cy5 [P] | 30 ng/L | [158] | |
| Okadaic acid (OA) | Optical | Direct | Mouse monoclonal anti-OA-HRP [P] | 0.1 μg/L | [159] |
| Amperometric | Direct | Mouse monoclonal anti-OA-AP [P] | 1.5 μg/L | [151] | |
| Piezoelectric | Capture | Monoclonal anti-OA [C] | 3.6 μg/L | [160] | |
| Amperometric | Capture | Monoclonal anti-OA [C] | 2 μg/L | [161] | |
| Amperometric | Indirect | Mouse monoclonal anti-OA [P]; goat anti-mouse-HRP or AP [D] | 0.03 μg/L | [162] | |
| Saxitoxin (STX) | Amperometric | Direct | Donkey anti-STX-glucose oxidase [P] | 2 μg/L | [150] |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Byrne, B.; Stack, E.; Gilmartin, N.; O’Kennedy, R. Antibody-Based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins. Sensors 2009, 9, 4407-4445. https://doi.org/10.3390/s90604407
Byrne B, Stack E, Gilmartin N, O’Kennedy R. Antibody-Based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins. Sensors. 2009; 9(6):4407-4445. https://doi.org/10.3390/s90604407
Chicago/Turabian StyleByrne, Barry, Edwina Stack, Niamh Gilmartin, and Richard O’Kennedy. 2009. "Antibody-Based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins" Sensors 9, no. 6: 4407-4445. https://doi.org/10.3390/s90604407
APA StyleByrne, B., Stack, E., Gilmartin, N., & O’Kennedy, R. (2009). Antibody-Based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins. Sensors, 9(6), 4407-4445. https://doi.org/10.3390/s90604407




