Bioinspired Designs, Molecular Premise and Tools for Evaluating the Ecological Importance of Antimicrobial Peptides
Abstract
:1. Introduction
2. Diverse Sources of AMPs
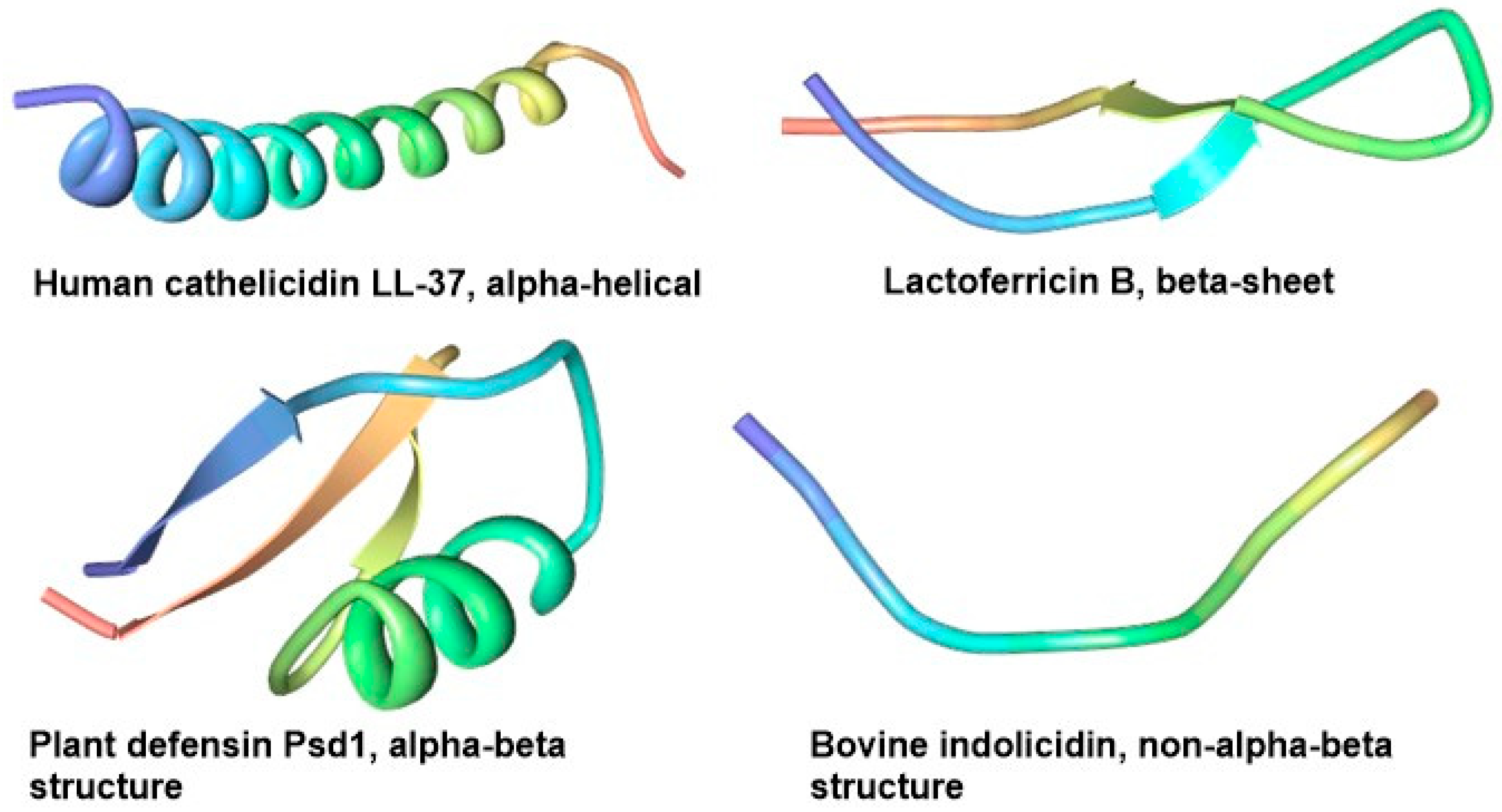
2.1. Structural Diversity of AMPs from Non-Bacterial Sources
2.2. Structural Diversity of AMPs from Bacterial Sources
3. General and Specific Modes of Action of AMPs
4. Potential Applications and Accompanying Challenges
5. The Paradox Surrounding Artificial AMPs and Semisynthetic Derivatives
6. Controlling the Unspecificity of AMPs
7. Molecular Premise for Evaluating the Ecotoxicity of AMPs
7.1. Cell Wall
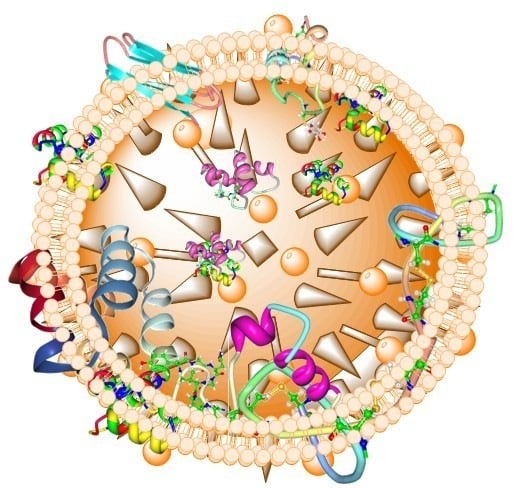
7.2. Lipid Bilayer Membrane and Pore Formation
7.3. Nucleic Acid Binding or Damage
7.4. Membrane Transporters and Receptors
7.5. Chaperones
7.6. RNA polymerase and 70S Ribosomes
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.; Ortiz, C.; Guzman, F.; Fernandez-Lafuente, R.; Torres, R. Antimicrobial peptides: Promising compounds against pathogenic microorganisms. Curr. Med. Chem. 2014, 21, 2299–2321. [Google Scholar] [CrossRef] [PubMed]
- Van Compernolle, S.; Smith, P.B.; Bowie, J.H.; Tyler, M.J.; Unutmaz, D.; Rollins-Smith, L.A. Inhibition of HIV infection by caerin 1 antimicrobial peptides. Peptides 2015, 71, 296–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delattin, N.; De Brucker, K.; De Cremer, K.; Cammue, B.; Thevissen, K. Antimicrobial peptides as a strategy to combat fungal biofilms. Curr. Top. Med. Chem. 2017, 17, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. 2013, 4, 294. [Google Scholar]
- Koopmans, T.; Wood, T.M.; Hart, P.; Kleijn, L.H.; Hendrickx, A.P.; Willems, R.J.; Breukink, E.; Martin, N.I. Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent compound. J. Am. Chem. Soc. 2015, 137, 9382–9389. [Google Scholar] [CrossRef] [PubMed]
- Baumann, T.; Nickling, J.H.; Bartholomae, M.; Buivydas, A.; Kuipers, O.P.; Budisa, N. Prospects of in vivo incorporation of noncanonical amino acids for the chemical diversification of antimicrobial peptides. Front. Microbiol. 2017, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Omardien, S.; Brul, S.; Zaat, S.A. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: Current progress made in understanding the mode of action and the response of bacteria. Front. Cell Dev. Biol. 2016, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B 2016, 371, 20150292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjos, M.; Oppegård, C.; Diep, D.B.; Nes, I.F.; Veening, J.W.; Nissen-Meyer, J.; Kristensen, T. Sensitivity to the two-peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell-wall synthesis. Mol. Microbiol. 2014, 92, 1177–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Kar, R.K.; Jana, J.; Saha, A.; Jana, B.; Krishnamoorthy, J.; Kumar, D.; Ghosh, S.; Chatterjee, S.; Bhunia, A. Indolicidin targets duplex DNA: Structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem 2014, 9, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Bera, S.; Shai, Y.; Mangoni, M.L.; Bhunia, A. NMR structure and binding of esculentin-1a (1–21) NH 2 and its diastereomer to lipopolysaccharide: Correlation with biological functions. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Miljkovic, M.; Uzelac, G.; Mirkovic, N.; Devescovi, G.; Diep, D.B.; Venturi, V.; Kojic, M. LsbB bacteriocin interacts with the third transmembrane domain of the YvjB receptor. Appl. Environ. Microbiol. 2016, 82, 5364–5374. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.S.; Ng, T.B. Northeast red beans produce a thermostable and pH-stable defensin-like peptide with potent antifungal activity. Cell Biochem. Biophys. 2013, 66, 637–648. [Google Scholar] [CrossRef] [PubMed]
- An, M.-Y.; Gao, J.; Zhao, X.-F.; Wang, J.-X. A new subfamily of penaeidin with an additional serine-rich region from kuruma shrimp (Marsupenaeus japonicus) contributes to antimicrobial and phagocytic activities. Dev. Comp. Immunol. 2016, 59, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Ebbensgaard, A.; Mordhorst, H.; Overgaard, M.T.; Nielsen, C.G.; Aarestrup, F.M.; Hansen, E.B. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS ONE 2015, 10, e0144611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.; Guo, H.; Ou, Y.; Liu, G.; Fang, X.; Liao, Z.; Ke, C.; Chen, Y.; Zhao, L.; Cao, Y. Purification and characterization of bacteriocin F1, a novel bacteriocin produced by Lactobacillus paracasei subsp. tolerans FX-6 from Tibetan kefir, a traditional fermented milk from Tibet. China. Food Control 2014, 42, 48–53. [Google Scholar]
- Svenson, J.; Stensen, W.; Brandsdal, B.-O.; Haug, B.E.; Monrad, J.; Svendsen, J.S. Antimicrobial peptides with stability toward tryptic degradation. Biochemistry 2008, 47, 3777–3788. [Google Scholar] [CrossRef] [PubMed]
- Greber, K.E.; Dawgul, M. Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 2017, 17, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Masuda, K.; Utsunomiya, R.; Shiraishi, K.; Mori, H.; Tohyama, M.; Sayama, K. TLN-58, newly discovered hCAP18 processing form found in the lesion vesicle of palmoplantar pustulosis. J. Dermatol. Sci. 2016, 84, e116. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Oyston, P.; Fox, M.; Richards, S.; Clark, G. Novel peptide therapeutics for treatment of infections. J. Med. Microbiol. 2009, 58, 977–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Slootweg, J.C.; Liskamp, R.M.; Rijkers, D.T. Scalable purification of the lantibiotic nisin and isolation of chemical/enzymatic cleavage fragments suitable for semi-synthesis. J. Pept. Sci. 2013, 19, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Ongey, E.L.; Yassi, H.; Pflugmacher, S.; Neubauer, P. Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol. Lett. 2017, 39, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Ryu, C.H.; Jun, J.; Kim, S.M.; Jeong, C.H.; Jeun, S.-S. IL-8 enhances the angiogenic potential of human bone marrow mesenchymal stem cells by increasing vascular endothelial growth factor. Cell Biol. Int. 2014, 38, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Cotter, P.D.; Hill, C.; Ross, R. Bioengineering lantibiotics for therapeutic success. Front. Microbiol. 2015, 6, 1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escano, J.; Smith, L. Multipronged approach for engineering novel peptide analogues of existing lantibiotics. Expert Opin. Drug Discov. 2015, 10, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Cotter, P.D.; Ross, R.P.; Hill, C. Bioengineering of the model lantibiotic nisin. Bioengineered 2015, 6, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapsch, K.; Bier, F.F.; Tadros, M.; von Nickisch-Rosenegk, M. Identification of antimicrobial peptides and immobilization strategy suitable for a covalent surface coating with biocompatible properties. Bioconjug. Chem. 2014, 25, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Ghosh, A.; Sharma, S.; Debnath, T.; Giri, B.; Bhunia, A. Probing the role of proline in the antimicrobial activity and lipopolysaccharide binding of indolicidin. J. Colloid Interface Sci. 2015, 452, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Leber, R.; Schmuck, M.; Kriechbaum, M.; Cordfunke, R.A.; Drijfhout, J.W.; de Breij, A.; Nibbering, P.H.; Kolb, D.; Lohner, K. Phospholipid-driven differences determine the action of the synthetic antimicrobial peptide OP-145 on Gram-positive bacterial and mammalian membrane model systems. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 2437–2447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.F.; Abdelkhalek, A.; Seleem, M.N. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016, 6, 29707. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Aboye, T.L.; Strömstedt, A.A.; Gunasekera, S.; Bruhn, J.G.; El-Seedi, H.; Rosengren, K.J.; Göransson, U. A Cactus-Derived Toxin-Like Cystine Knot Peptide with Selective Antimicrobial Activity. ChemBioChem 2015, 16, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Dueñas-Cuellar, R.A.; Kushmerick, C.; Naves, L.A.; Batista, I.F.; Guerrero-Vargas, J.A.; Pires, O.R., Jr.; Fontes, W.; Castro, M.S. Cm38: A new antimicrobial peptide active against Klebsiella pneumoniae is homologous to Cn11. Protein Pep. Lett. 2015, 22, 164–172. [Google Scholar]
- Hassan, M.; Kjos, M.; Nes, I.; Diep, D.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Avitabile, C.; Capparelli, R.; Rigano, M.; Fulgione, A.; Barone, A.; Pedone, C.; Romanelli, A. Antimicrobial peptides from plants: Stabilization of the γ core of a tomato defensin by intramolecular disulfide bond. J. Pept. Sci. 2013, 19, 240–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, V.; Underhill, A.; Baber, I.; Sylla, L.; Baby, M.; Larget-Thiery, I.; Zettor, A.; Bourgouin, C.; Langel, Ü.; Faye, I. Killer bee molecules: Antimicrobial peptides as effector molecules to target sporogonic stages of Plasmodium. PLoS Pathog. 2013, 9, e1003790. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I. Evolution of antimicrobial peptides: A view from the cystine chapel. In Antimicrobial Peptides and Innate Immunity; Springer: New Yok, NY, USA, 2013; pp. 1–27. [Google Scholar]
- Boman, H. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Guaní-Guerra, E.; Santos-Mendoza, T.; Lugo-Reyes, S.O.; Terán, L.M. Antimicrobial peptides: General overview and clinical implications in human health and disease. Clin. Immunol. 2010, 135, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Mattoo, A.K. Plant antimicrobial peptides. In Host Defense Peptides and Their Potential as Therapeutic Agents; Springer: New Yok, NY, USA, 2016; pp. 111–136. [Google Scholar]
- Bhat, Z.; Kumar, S.; Bhat, H.F. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef] [PubMed]
- Kamali Alamdari, E.; Ehsani, M. Antimicrobial peptides derived from milk: A review. J. Food Biosci. Technol. 2017, 7, 49–56. [Google Scholar]
- Théolier, J.; Fliss, I.; Jean, J.; Hammami, R. MilkAMP: A comprehensive database of antimicrobial peptides of dairy origin. Dairy Sci. Technol. 2014, 94, 181–193. [Google Scholar] [CrossRef]
- Besse, A.; Peduzzi, J.; Rebuffat, S.; Carre-Mlouka, A. Antimicrobial peptides and proteins in the face of extremes: Lessons from archaeocins. Biochimie 2015, 118, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Bai, L.; Zhu, L.; Yang, L.; Zhang, X. Marine algae-derived bioactive peptides for human nutrition and health. J. Agric. Food Chem. 2014, 62, 9211–9222. [Google Scholar] [CrossRef] [PubMed]
- Matejuk, A.; Leng, Q.; Begum, M.; Woodle, M.; Scaria, P.; Chou, S.; Mixson, A. Peptide-based antifungal therapies against emerging infections. Drugs Future 2010, 35, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Mansour, S.C.; Hancock, R.E. Antimicrobial Peptides: An Introduction. Methods Mol. Biol. 2017, 1548, 3–22. [Google Scholar] [PubMed]
- Wang, G.; Mishra, B.; Lau, K.; Lushnikova, T.; Golla, R.; Wang, X. Antimicrobial peptides in 2014. Pharmaceuticals 2015, 8, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Solstad, R.G.; Li, C.; Isaksson, J.; Johansen, J.; Svenson, J.; Stensvåg, K.; Haug, T. Novel Antimicrobial Peptides EeCentrocins 1, 2 and EeStrongylocin 2 from the Edible Sea Urchin Echinus esculentus Have 6-Br-Trp Post-Translational Modifications. PLoS ONE 2016, 11, e0151820. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Yomogida, S.; Nagaoka, I.; Yamashita, T. Purification of the 11-and 5-kDa antibacterial polypeptides from guinea pig neutrophils. Arch. Biochem. Biophys. 1996, 328, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr. Pharm. Des. 2002, 8, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.P.; Kariolis, M.S.; Cochran, J.R. Cystine-knot peptides engineered with specificities for αIIbβ3 or αIIbβ3 and αvβ3 integrins are potent inhibitors of platelet aggregation. J. Mol. Recognit. 2011, 24, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Treggiari, D.; Dalbeni, A.; Minuz, P.; Pandolfini, T. Plant cystine-knot peptides: Pharmacological perspectives. Br. J. Clin. Pharmacol. 2017, 83, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.; Tang, M.; Wu, X.; Buffy, J.; Waring, A.; Sherman, M.; Hong, M. Membrane-bound dimer structure of a β-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry 2006, 45, 8341–8349. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E. Cystine knot growth factors and their functionally versatile proregions. Biol. Chem. 2017, 398, 1295–1308. [Google Scholar] [CrossRef] [PubMed]
- Pallaghy, P.K.; Norton, R.S.; Nielsen, K.J.; Craik, D.J. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiche, L.; Heitz, A.; Gelly, J.-C.; Gracy, J.; Chau, P.T.; Ha, P.T.; Hernandez, J.-F.; Le-Nguyen, D. Squash inhibitors: From structural motifs to macrocyclic knottins. Curr. Protein Pept. Sci. 2004, 5, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Daly, N.L.; Craik, D.J. Bioactive cystine knot proteins. Curr. Opin. Chem. Biol. 2011, 15, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Göransson, U.; Burman, R.; Gunasekera, S.; Strömstedt, A.A.; Rosengren, K.J. Circular proteins from plants and fungi. J. Biol. Chem. 2012, 287, 27001–27006. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Ke, T.; Liu, R.; Yu, J.; Dong, C.; Cheng, M.; Huang, J.; Liu, S. Identification of a novel proline-rich antimicrobial peptide from Brassica napus. PLoS ONE 2015, 10, e0137414. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: Converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Rucker, A.L.; Creamer, T.P. Polyproline II helical structure in protein unfolded states: Lysine peptides revisited. Protein Sci. 2002, 11, 980–985. [Google Scholar] [PubMed]
- Guzmán, F.; Marshall, S.; Ojeda, C.; Albericio, F.; Carvajal-Rondanelli, P. Inhibitory effect of short cationic homopeptides against Gram-positive bacteria. J. Pept. Sci. 2013, 19, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Bieler, S.; Silva, F.; Soto, C.; Belin, D. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 2006, 188, 7049–7061. [Google Scholar] [CrossRef] [PubMed]
- Cabiaux, V.; Agerberths, B.; Johansson, J.; Homble, F.; Goormaghtigh, E.; Ruysschaert, J.M. Secondary structure and membrane interaction of PR-39, a Pro+ Arg-rich antibacterial peptide. Eur. J. Biochem. 1994, 224, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Raj, P.A.; Edgerton, M. Functional domain and poly-l-proline II conformation for candidacidal activity of bactenecin 5. FEBS Lett. 1995, 368, 526–530. [Google Scholar] [PubMed]
- Kuriakose, J.; Hernandez-Gordillo, V.; Nepal, M.; Brezden, A.; Pozzi, V.; Seleem, M.N.; Chmielewski, J. Targeting intracellular pathogenic bacteria with unnaturalproline-rich peptides: Coupling antibacterial activity with macrophage penetration. Angew. Chem. 2013, 125, 9846–9849. [Google Scholar] [CrossRef]
- Nepal, M.; Thangamani, S.; Seleem, M.N.; Chmielewski, J. Targeting intracellular bacteria with an extended cationic amphiphilic polyproline helix. Org. Biomol. Chem. 2015, 13, 5930–5936. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Rondanelli, P.; Aróstica, M.; Marshall, S.H.; Albericio, F.; Álvarez, C.A.; Ojeda, C.; Aguilar, L.F.; Guzmán, F. Inhibitory effect of short cationic homopeptides against Gram-negative bacteria. Amino Acids 2016, 48, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Rios, A.C.; Moutinho, C.G.; Pinto, F.C.; Del Fiol, F.S.; Jozala, A.; Chaud, M.V.; Vila, M.M.D.C.; Teixeira, J.A.; Balcão, V.M. Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol. Res. 2016, 191, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.L.; Kennedy, A.M.; Spuches, A.M.; Venugopal, D.; Bhonsle, J.B.; Hicks, R.P. Spectroscopic and thermodynamic evidence for antimicrobial peptide membrane selectivity. Chem. Phys. Lipids 2010, 163, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, P.; Biondi, B.; Marchiani, A.; Antolini, N.; Calderan, A. Cell-penetrating peptides: A comparative study on lipid affinity and cargo delivery properties. Pharmaceuticals 2010, 3, 1045–1062. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, P.; Reichert, J.; Bürck, J.; Ulrich, A.S. Antimicrobial and cell-penetrating peptides induce lipid vesicle fusion by folding and aggregation. Eur. Biophys. J. 2012, 41, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Pires, J.; Siriwardena, T.N.; Stach, M.; Tinguely, R.; Kasraian, S.; Luzzaro, F.; Leib, S.L.; Darbre, T.; Reymond, J.-L.; Endimiani, A. In vitro activity of the novel antimicrobial peptide dendrimer G3KL against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 7915–7918. [Google Scholar] [CrossRef] [PubMed]
- Kumaresan, V.; Bhatt, P.; Ganesh, M.-R.; Harikrishnan, R.; Arasu, M.; Al-Dhabi, N.A.; Pasupuleti, M.; Marimuthu, K.; Arockiaraj, J. A novel antimicrobial peptide derived from fish goose type lysozyme disrupts the membrane of Salmonella enterica. Mol. Immunol. 2015, 68, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-L.; Yip, B.-S.; Chen, K.-H.; Yu, H.-Y.; Chih, Y.-H.; Cheng, H.-T.; Chou, Y.-T.; Cheng, J.-W. Novel antimicrobial peptides with high anticancer activity and selectivity. PLoS ONE 2015, 10, e0126390. [Google Scholar] [CrossRef] [PubMed]
- Marani, M.M.; Perez, L.O.; de Araujo, A.R.; Plácido, A.; Sousa, C.F.; Quelemes, P.V.; Oliveira, M.; Gomes-Alves, A.G.; Pueta, M.; Gameiro, P.; et al. Thaulin-1: The first antimicrobial peptide isolated from the skin of a Patagonian frog Pleurodema thaul (Anura: Leptodactylidae: Leiuperinae) with activity against Escherichia coli. Gene 2016, 605, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Hwang, J.-S.; Lee, D.G. A novel antimicrobial peptide, scolopendin, from Scolopendra subspinipes mutilans and its microbicidal mechanism. Biochimie 2015, 118, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; C Rea, M.; D Cotter, P.; Hill, C.; Paul Ross, R. The sactibiotic subclass of bacteriocins: An update. Curr. Protein Pept. Sci. 2015, 16, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015, 48, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ducasse, R.; Zirah, S.; Blond, A.; Goulard, C.; Lescop, E.; Giraud, C.; Hartke, A.; Guittet, E.; Pernodet, J.-L.; et al. Characterization of sviceucin from Streptomyces provides insights into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem.Biol. 2015, 10, 2641–2649. [Google Scholar] [CrossRef] [PubMed]
- Kersten, R.D.; Yang, Y.-L.; Xu, Y.; Cimermancic, P.; Nam, S.-J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maqueda, M.; Sánchez-Hidalgo, M.; Fernández, M.; Montalbán-López, M.; Valdivia, E.; Martínez-Bueno, M. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 2008, 32, 2–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalbán-López, M.; Sánchez-Hidalgo, M.; Cebrián, R.; Maqueda, M. Discovering the bacterial circular proteins: Bacteriocins, cyanobactins, and pilins. J. Biol. Chem. 2012, 287, 27007–27013. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; van Belkum, M.J.; Lohans, C.T.; McKay, R.T.; Miskolzie, M.; Vederas, J.C. Solution structure of acidocin B, a circular bacteriocin produced by Lactobacillus acidophilus M46. Appl. Environ. Microbiol. 2015, 81, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Himeno, K.; Rosengren, K.J.; Inoue, T.; Perez, R.H.; Colgrave, M.L.; Lee, H.S.; Chan, L.Y.; Henriques, S.T.; Fujita, K.; Ishibashi, N. Identification, characterization, and three-dimensional structure of the novel circular bacteriocin, enterocin NKR-5-3B, from Enterococcus faecium. Biochemistry 2015, 54, 4863–4876. [Google Scholar] [CrossRef] [PubMed]
- Martin-Visscher, L.A.; van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.H.; Chakicherla, A.; Hansen, J.N. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 1998, 273, 23134–23142. [Google Scholar] [CrossRef] [PubMed]
- Garcia De Gonzalo, C.V.; Zhu, L.; Oman, T.J.; Van Der Donk, W.A. NMR structure of the S-linked glycopeptide sublancin 168. ACS Chem. Biol. 2014, 9, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Babasaki, K.; Takao, T.; Shimonishi, Y.; Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985, 98, 585–603. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; O’Sullivan, O.; Rea, M.C.; Cotter, P.D.; Ross, R.P.; Hill, C. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS ONE 2011, 6, e20852. [Google Scholar] [CrossRef] [PubMed]
- Kawulka, K.; Sprules, T.; McKay, R.T.; Mercier, P.; Diaper, C.M.; Zuber, P.; Vederas, J.C. Structure of Subtilosin A, an Antimicrobial Peptide from Bacillus s ubtilis with Unusual Posttranslational Modifications Linking Cysteine Sulfurs to α-Carbons of Phenylalanine and Threonine. J. Am. Chem. Soc. 2003, 125, 4726–4727. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Lopez, M.; Sanchez-Hidalgo, M.; Valdivia, E.; Martinez-Bueno, M.; Maqueda, M. Are bacteriocins underexploited? Novel applications for old antimicrobials. Curr. Pharm. Biotechnol. 2011, 12, 1205–1220. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, F.; Lazzarini, A.; Carrano, L.; Corti, E.; Ciciliato, I.; Gastaldo, L.; Candiani, P.; Losi, D.; Marinelli, F.; Selva, E. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 2008, 15, 22–31. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.A.; Donia, M.S.; Schmidt, E.W. Ribosomal peptide natural products: Bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009, 26, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Dischinger, J.; Basi Chipalu, S.; Bierbaum, G. Lantibiotics: Promising candidates for future applications in health care. Int. J. Med. Microbiol. 2014, 304, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, F. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry 2007, 46, 5884–5895. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.C.S.; Iorio, M.; Monciardini, P.; Simone, M.; Brunati, C.; Gaspari, E.; Maffioli, S.I.; Wellington, E.; Sosio, M.; Donadio, S. Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J. Nat. Prod. 2015, 78, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.; Sasso, O.; Maffioli, S.I.; Bertorelli, R.; Monciardini, P.; Sosio, M.; Bonezzi, F.; Summa, M.; Brunati, C.; Bordoni, R. A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem. Biol. 2013, 9, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Rink, R.; Arkema-Meter, A.; Baudoin, I.; Post, E.; Kuipers, A.; Nelemans, S.; Akanbi, M.H.J.; Moll, G. To protect peptide pharmaceuticals against peptidases. J. Pharmacol. Toxicol. Methods 2010, 61, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Okesli, A.E.; van der Donk, W.A. Mechanistic studies of Ser/Thr dehydration catalyzed by a member of the LanL lanthionine synthetase family. Biochemistry 2011, 50, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; van der Donk, W.A. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa bacteriocins: Diversity and new developments. Int. J. Mol. Sci. 2012, 13, 16668–16707. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Novel bacteriocins from lactic acid bacteria (LAB): Various structures and applications. Microb. Cell Fact. 2014, 13, S3. [Google Scholar] [CrossRef] [PubMed]
- Sit, C.S.; Lohans, C.T.; van Belkum, M.J.; Campbell, C.D.; Miskolzie, M.; Vederas, J.C. Substitution of a conserved disulfide in the type IIa bacteriocin, leucocin A, with L-leucine and L-serine residues: Effects on activity and three-dimensional structure. ChemBioChem 2012, 13, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.L.; Lamosa, P.; Rodríguez, A.; Martínez, B. Structure and properties of the metastable bacteriocin Lcn972 from Lactococcus lactis. J. Mol. Struct. 2013, 1031, 207–210. [Google Scholar] [CrossRef]
- Azmi, F.; Skwarczynski, M.; Toth, I. Towards the development of synthetic antibiotics: Designs inspired by natural antimicrobial peptides. Curr. Med. Chem. 2016, 23, 4610–4624. [Google Scholar] [CrossRef] [PubMed]
- Khara, J.S.; Priestman, M.; Uhía, I.; Hamilton, M.S.; Krishnan, N.; Wang, Y.; Yang, Y.Y.; Langford, P.R.; Newton, S.M.; Robertson, B.D. Unnatural amino acid analogues of membrane-active helical peptides with anti-mycobacterial activity and improved stability. J. Antimicrob. Chemother. 2016, 71, 2181–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Xu, B. Inspiration from the mirror: d-amino acid containing peptides in biomedical approaches. Biomol. Concepts 2016, 7, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.-G.; Shai, Y. The consequence of sequence alteration of an amphipathic α-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.d.S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. 2013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, B.; Gorr, S.-U. Antimicrobial peptides mechanisms of action and resistance. J. Dent. Res. 2016, 6, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Freund, S.; Jung, G.; Gutbrod, O.; Foikers, G.; Gibbons, W.A.; Allgaier, H.; Werner, R. The solution structure of the lantibiotic gallidermin. Biopolymers 1991, 31, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-helical antimicrobial peptides. Pept. Sci. 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Sánchez-Barrena, M.; Martınez-Ripoll, M.; Gálvez, A.; Valdivia, E.; Maqueda, M.; Cruz, V.; Albert, A. Structure of bacteriocin AS-48: From soluble state to membrane bound state. J. Mol. Biol. 2003, 334, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta (BBA)-Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Makin, S.A.; Beveridge, T.J. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology 1996, 142, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Lohner, K. Membrane-active antimicrobial peptides as template structures for novel antibiotic agents. Curr. Top. Med. Chem. 2017, 17, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr. Antibacterial peptides isolated from insects. J. Pept. Sci. 2000, 6, 497–511. [Google Scholar] [CrossRef]
- Gennaro, R.; Zanetti, M.; Benincasa, M.; Podda, E.; Miani, M. Pro-rich antimicrobial peptides from animals: Structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002, 8, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Avitabile, C.; D’Andrea, L.D.; Romanelli, A. Circular dichroism studies on the interactions of antimicrobial peptides with bacterial cells. Sci. Rep. 2014, 4, 4293. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, C.; Brede, D.A.; Hernández, P.E.; Nes, I.F.; Diep, D.B. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother. 2012, 56, 2908–2915. [Google Scholar] [CrossRef] [PubMed]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzelac, G.; Kojic, M.; Lozo, J.; Aleksandrzak-Piekarczyk, T.; Gabrielsen, C.; Kristensen, T.; Nes, I.F.; Diep, D.B.; Topisirovic, L. A Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. 2013, 195, 5614–5621. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Grzela, R.; Giglione, C.; Meinnel, T.; Gennaro, R.; Mergaert, P.; Scocchi, M. The host antimicrobial peptide Bac7 1-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 2014, 21, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Krizsan, A.; Volke, D.; Weinert, S.; Sträter, N.; Knappe, D.; Hoffmann, R. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew. Chem. Int. Ed. 2014, 53, 12236–12239. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, J.; Sineva, E.; Knight, J.; Levy, R.M.; Ebright, R.H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 2004, 14, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Kuznedelov, K.; Semenova, E.; Knappe, T.A.; Mukhamedyarov, D.; Srivastava, A.; Chatterjee, S.; Ebright, R.H.; Marahiel, M.A.; Severinov, K. The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J. Mol. Biol. 2011, 412, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Lüthy, C.; Decarli, P.; Mignogna, G.; Christen, P.; Gennaro, R. The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Ther. 2009, 15, 147–155. [Google Scholar] [CrossRef]
- Wilmes, M.; Stockem, M.; Bierbaum, G.; Schlag, M.; Götz, F.; Tran, D.Q.; Schaal, J.B.; Ouellette, A.J.; Selsted, M.E.; Sahl, H.-G. Killing of staphylococci by θ-defensins involves membrane impairment and activation of autolytic enzymes. Antibiotics 2014, 3, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, G.; Sahl, H.G. Lantibiotics: Mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, I.; Hirota, S.; Niyonsaba, F.; Hirata, M.; Adachi, Y.; Tamura, H.; Tanaka, S.; Heumann, D. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobial peptides by replacement with hydrophobic and cationic amino acid residues. Clin. Diagn. Lab. Immunol. 2002, 9, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Bociek, K.; Ferluga, S.; Mardirossian, M.; Benincasa, M.; Tossi, A.; Gennaro, R.; Scocchi, M. Lipopolysaccharide phosphorylation by the waay kinase affects the susceptibility of Escherichia coli to the human antimicrobial peptide LL-37. J. Biol. Chem. 2015, 290, 19933–19941. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Masuda, R.; Dazai, Y.; Mima, T.; Koide, T. Structure–Activity Relationships and Action Mechanisms of Collagen-like Antimicrobial Peptides. Pept. Sci. 2017, 108. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Zhou, J.; Liu, G.; Chen, F.; Chen, Y.; Gao, X.; Dixon, W.; Song, M.; Xiao, H.; Cao, Y. Membrane disruption and DNA binding of Staphylococcus aureus cell induced by a novel antimicrobial peptide produced by Lactobacillus paracasei subsp. tolerans FX-6. Food Control 2016, 59, 609–613. [Google Scholar] [CrossRef]
- Costa, F.; Carvalho, I.F.; Montelaro, R.C.; Gomes, P.; Martins, M.C.L. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 2011, 7, 1431–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosikowska, P.; Lesner, A. Antimicrobial peptides (AMPs) as drug candidates: A patent review (2003–2015). Expert Opin. Ther. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Strempel, N.; Strehmel, J.; Overhage, J. Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr. Pharm. Des. 2015, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordström, R.; Malmsten, M. Delivery systems for antimicrobial peptides. Adv. Colloid Interface Sci. 2017, 242, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Leake, I. IBD: Cathelicidin can reverse intestinal fibrosis in models of colitis. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr.; Ostorhazi, E. Therapeutic utility of antibacterial peptides in wound healing. Expert Rev. Anti Infect. Ther. 2015, 13, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.; Olívia Pereira, M. Mini-review: Antimicrobial peptides and enzymes as promising candidates to functionalize biomaterial surfaces. Biofouling 2014, 30, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Jiang, Y.; Nüsslein, K.; Rotello, V.M.; Santore, M.M. Antimicrobial surfaces containing cationic nanoparticles: How immobilized, clustered, and protruding cationic charge presentation affects killing activity and kinetics. Colloids Surf. B. Biointerfaces 2015, 125, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; MacDougall, C.; Lim, M. Antimicrobial surfaces to prevent healthcare-associated infections: A systematic review. J. Hosp. Infect. 2016, 92, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Salwiczek, M.; Qu, Y.; Gardiner, J.; Strugnell, R.A.; Lithgow, T.; McLean, K.M.; Thissen, H. Emerging rules for effective antimicrobial coatings. Trends Biotechnol. 2014, 32, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Lillehoj, P.B.; Kaplan, C.W.; He, J.; Shi, W.; Ho, C.-M. Rapid, electrical impedance detection of bacterial pathogens using immobilized antimicrobial peptides. J. Lab. Autom. 2014, 19, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Onaizi, S.A.; Leong, S.S. Tethering antimicrobial peptides: Current status and potential challenges. Biotechnol. Adv. 2011, 29, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; Beyermann, M.; Dathe, M. Immobilization reduces the activity of surface-bound cationic antimicrobial peptides with no influence upon the activity spectrum. Antimicrob. Agents Chemother. 2009, 53, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Shalev, T.; Gopin, A.; Bauer, M.; Stark, R.W.; Rahimipour, S. Non-leaching antimicrobial surfaces through polydopamine bio-inspired coating of quaternary ammonium salts or an ultrashort antimicrobial lipopeptide. J. Mater. Chem. 2012, 22, 2026–2032. [Google Scholar] [CrossRef] [Green Version]
- Murata, H.; Koepsel, R.R.; Matyjaszewski, K.; Russell, A.J. Permanent, non-leaching antibacterial surfaces—2: How high density cationic surfaces kill bacterial cells. Biomaterials 2007, 28, 4870–4879. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Zumbuehl, A. Non-leaching surfaces capable of killing microorganisms on contact. J. Mater. Chem. 2009, 19, 7796–7806. [Google Scholar] [CrossRef]
- Goodwin, D.; Simerska, P.; Toth, I. Peptides as therapeutics with enhanced bioactivity. Curr. Med. Chem. 2012, 19, 4451–4461. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.J.; Appleyard, A.N.; Bargallo, J.C.; Wadman, S.N. Actagardine Derivatives, and Pharmaceutical Use Thereof. U.S. Patent WO 2,010,082,019 A1, 22 July 2010. [Google Scholar]
- Boakes, S.; Weiss, W.J.; Vinson, M.; Wadman, S.; Dawson, M.J. Antibacterial activity of the novel semisynthetic lantibiotic NVB333 in vitro and in experimental infection models. J. Antibiot. 2016, 69, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, D.C.; Lee, J.J.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007, 2, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Cox, V.E.; Gaucher, E.A. Molecular evolution directs protein translation using unnatural amino acids. Curr. Protoc. Chem. Biol. 2015, 223–228. [Google Scholar]
- Sadler, K.; Tam, J.P. Peptide dendrimers: Applications and synthesis. Rev. Mol. Biotechnol. 2002, 90, 195–229. [Google Scholar] [CrossRef]
- Pini, A.; Falciani, C.; Bracci, L. Branched peptides as therapeutics. Curr. Protein Pept. Sci. 2008, 9, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Dulcey, A.E.; Balagurusamy, V.S.; Miura, Y.; Smidrkal, J.; Peterca, M.; Nummelin, S.; Edlund, U.; Hudson, S.D.; Heiney, P.A. Self-assembly of amphiphilic dendritic dipeptides into helical pores. Nature 2004, 430, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringstad, L.; Schmidtchen, A.; Malmsten, M. Effects of single amino acid substitutions on peptide interaction with lipid membranes and bacteria–variants of GKE21, an internal sequence from human LL-37. Colloids Surf. Physicochem. Eng. Asp. 2010, 354, 65–71. [Google Scholar] [CrossRef]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Interactions of tryptophan-rich cathelicidin antimicrobial peptides with model membranes studied by differential scanning calorimetry. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 2447–2458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic α helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, M.; Runti, G.; Mardirossian, M.; Gennaro, R.; Scocchi, M. Methods for elucidating the mechanism of action of proline-rich and other non-lytic antimicrobial peptides. Methods Mol. Biol. 2017, 1548, 283–295. [Google Scholar] [PubMed]
- Xhindoli, D.; Pacor, S.; Guida, F.; Antcheva, N.; Tossi, A. Native oligomerization determines the mode of action and biological activities of human cathelicidin LL-37. Biochem. J. 2014, 457, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.; Gudmundsson, G.H.; Rottenberg, M.E.; Berndt, K.D.; Agerberth, B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 1998, 273, 3718–3724. [Google Scholar] [CrossRef] [PubMed]
- Zelezetsky, I.; Pacor, S.; Pag, U.; Papo, N.; Shai, Y.; Sahl, H.-G.; Tossi, A. Controlled alteration of the shape and conformational stability of α-helical cell-lytic peptides: Effect on mode of action and cell specificity. Biochem. J. 2005, 390, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, M.; Walse, B.R.; Svensson, B.; Malmsten, M.; Schmidtchen, A. Rational design of antimicrobial C3a analogues with enhanced effects against Staphylococci using an integrated structure and function-based approach. Biochemistry 2008, 47, 9057–9070. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Zuckermann, R.N.; Jenssen, H. Structure-activity relationship study of novel peptoids that mimic the structure of antimicrobial peptides. Antimicrob. Agents Chemother. 2015, 59, 4112–4120. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Cheng, J.; Liang, Z.C.; Xu, L.; Lou, W.; Bao, C.; Ong, Z.Y.; Dong, H.; Yang, Y.Y.; Fan, W. Short synthetic β-sheet antimicrobial peptides for the treatment of multidrug-resistant Pseudomonas aeruginosa burn wound infections. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Vasil, A.I.; Gera, L.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides to Target Gram-negative Pathogens, Acinetobacter baumannii and Pseudomonas aeruginosa: Utilization of Charge,‘Specificity Determinants’, Total Hydrophobicity, Hydrophobe Type and Location as Design Parameters to Improve the Therapeutic Ratio. Chem. Biol. Drug Des. 2011, 77, 225–240. [Google Scholar] [PubMed]
- Chu, H.-L.; Yu, H.-Y.; Yip, B.-S.; Chih, Y.-H.; Liang, C.-W.; Cheng, H.-T.; Cheng, J.-W. Boosting salt resistance of short antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 4050–4052. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hanke, M.L.; Mishra, B.; Lushnikova, T.; Heim, C.E.; Chittezham Thomas, V.; Bayles, K.W.; Kielian, T. Transformation of human cathelicidin LL-37 into selective, stable, and potent antimicrobial compounds. ACS Chem. Biol. 2014, 9, 1997–2002. [Google Scholar] [CrossRef] [PubMed]
- Naghmouchi, K.; Le Lay, C.; Baah, J.; Drider, D. Antibiotic and antimicrobial peptide combinations: Synergistic inhibition of Pseudomonas fluorescens and antibiotic-resistant variants. Res. Microbiol. 2012, 163, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Nuding, S.; Frasch, T.; Schaller, M.; Stange, E.F.; Zabel, L.T. Synergistic effects of antimicrobial peptides and antibiotics against Clostridium difficile. Antimicrob. Agents Chemother. 2014, 58, 5719–5725. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.; Handy, R.D.; Garrod, J.; Owen, R. Ecotoxicity test methods and environmental hazard assessment for engineered nanoparticles. Ecotoxicology 2008, 17, 421. [Google Scholar] [CrossRef] [PubMed]
- Bour, A.; Mouchet, F.; Silvestre, J.; Gauthier, L.; Pinelli, E. Environmentally relevant approaches to assess nanoparticles ecotoxicity: A review. J. Hazard. Mater. 2015, 283, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yan, J.; Dang, W.; Liu, X.; Chen, R.; Zhang, J.; Zhang, B.; Zhang, W.; Kai, M.; Yan, W. Membrane active antimicrobial activity and molecular dynamics study of a novel cationic antimicrobial peptide polybia-MPI, from the venom of Polybia paulista. Peptides 2013, 39, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kim, H.K.; Am Kim, S.; Park, Y.; Park, S.-C.; Jang, S.-H.; Hahm, K.-S. Fungicidal effect of indolicidin and its interaction with phospholipid membranes. Biochem. Biophys. Res. Commun. 2003, 305, 305–310. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Musielak, T.J.; Schenkel, L.; Kolb, M.; Henschen, A.; Bayer, M. A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod. 2015, 28, 161–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartnicki-Garcia, S.; Nickerson, W.J. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim. Biophys. Acta 1962, 58, 102–119. [Google Scholar] [CrossRef]
- Los, F.C.; Randis, T.M.; Aroian, R.V.; Ratner, A.J. Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol. Rev. 2013, 77, 173–207. [Google Scholar] [CrossRef] [PubMed]
- Dal Peraro, M.; Van Der Goot, F.G. Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Bacalum, M.; Radu, M. Cationic antimicrobial peptides cytotoxicity on mammalian cells: An analysis using therapeutic index integrative concept. Int. J. Pept. Res. Ther. 2015, 21, 47–55. [Google Scholar] [CrossRef]
- Slavík, J. Anilinonaphthalene sulfonate as a probe of membrane composition and function. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1982, 694, 1–25. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Thennarasu, S.; Tan, A.; Lee, D.-K.; Clayberger, C.; Krensky, A.M. Cell selectivity correlates with membrane-specific interactions: A case study on the antimicrobial peptide G15 derived from granulysin. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 154–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Yang, Z.; Weisshaar, J.C. Single-cell, real-time detection of oxidative stress induced in Escherichia coli by the antimicrobial peptide CM15. Proc. Natl. Acad. Sci. USA 2015, 112, E303–E310. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants–from models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.J.; Troxler, R.F.; Oppenheim, F.G. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc. Natl. Acad. Sci. USA 2001, 98, 14637–14642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pflugmacher, S.; Kwon, K.-S.; Baik, S.; Kim, S.; Kühn, S.; Esterhuizen-Londt, M. Physiological responses of Cladophora glomerata to cyanotoxins: A potential new phytoremediation species for the Green Liver Systems. Toxicol. Environ. Chem. 2016, 98, 241–259. [Google Scholar] [CrossRef]
- Vilvert, E.; Contardo-Jara, V.; Esterhuizen-Londt, M.; Pflugmacher, S. The effect of oxytetracycline on physiological and enzymatic defense responses in aquatic plant species Egeria densa, Azolla caroliniana, and Taxiphyllum barbieri. Toxicol. Environ. Chem. 2017, 99, 104–116. [Google Scholar] [CrossRef]
- Pflugmacher, S. Promotion of oxidative stress in the aquatic macrophyte Ceratophyllum demersum during biotransformation of the cyanobacterial toxin microcystin-LR. Aquat. Toxicol. 2004, 70, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Anderson, D. The Comet Assay in Toxicology; Royal Society of Chemistry: Cambridge, UK, 2016; Volume 30. [Google Scholar]
- de Lapuente, J.; Lourenço, J.; Mendo, S.A.; Borràs, M.; Martins, M.G.; Costa, P.M.; Pacheco, M. The Comet assay and its applications in the field of ecotoxicology: A mature tool that continues to expand its perspectives. Front. Genet. 2015, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Frenzilli, G.; Bean, T.; Lyons, B. The application of the Comet assay in aquatic environments. Comet Assay Toxicol. 2016, 30, 354. [Google Scholar]
- Cotter, P.D. An ‘Upp’-turn in bacteriocin receptor identification. Mol. Microbiol. 2014, 92, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, G.; Atallah, M.; Morolli, B.; Calléja, F.; Ras-Verloop, N.; Huijskens, I.; Raamsman, M.; van de Water, B.; Vrieling, H. The ToxTracker assay: Novel GFP reporter systems that provide mechanistic insight into the genotoxic properties of chemicals. Toxicol. Sci. 2011, 125, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, G.; Derr, R.S.; Misovic, B.; Morolli, B.; Calléja, F.M.; Vrieling, H. The extended ToxTracker assay discriminates between induction of DNA damage, oxidative stress, and protein misfolding. Toxicol. Sci. 2015, 150, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr.; O, I.; Rogers, M.E.; Consolvo, P.J.; Condie, B.A.; Lovas, S.; Bulet, P.; Blaszczyk-Thurin, M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar] [CrossRef] [PubMed]
- Mashaghi, A.; Bezrukavnikov, S.; Minde, D.P.; Wentink, A.S.; Kityk, R.; Zachmann-Brand, B.; Mayer, M.P.; Kramer, G.; Bukau, B.; Tans, S.J. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 2016, 539, 448–451. [Google Scholar] [CrossRef] [PubMed]




| Target Molecule | Killing Method | Examples | Reference |
|---|---|---|---|
| MLT | Efflux of intracellular and/or influx of extracellular solutes | Garvicin ML | [140] |
| Man-PTS | Efflux of intracellular and/or influx of extracellular solutes | Pediocin-like bacteriocins, Lactococcin A, microcin E492 | [73,141] |
| UppP | Disrupts cell-wall synthesis | Lactococcin G and Enterocin 1071 | [10] |
| MBM | Prevent proteolytic breakdown of a misfolded protein | LsbB | [13,142] |
| 70S ribosome | Inhibit protein synthesis | Bac7(1–35), insect-derived PrAMPs | [143,144] |
| RPol | Inhibit transcription by obstructing RNA polymerase activity | Microcin J25, Capistruin | [145,146] |
| DnaK | Inhibit ATPase activity | Pyrrhocoricin, Bac7 (PrAMPs) | [137,147,148] |
| LTA | Release of autolysin | RTD2 and Pep5 | [149] |
| Lipid II | Inhibit cell wall biosynthesis, pore formation | nisin | [150] |
| LPS | Restrict LPS binding to CD14+ cells and hence prevent fatal septic shock syndrome Interact with AMP and enable folding | Human cathelicidin LL-37 Indolicidin, magainin 2, cecropin A, esculentin-derived AMP, pyrrhocoricin | [12,33,137,139,151,152] |
| DNA/RNA | Inhibiting DNA replication | Indolicidin, Buforin II, tachyplesin, RR4 | [11,153,154] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ongey, E.L.; Pflugmacher, S.; Neubauer, P. Bioinspired Designs, Molecular Premise and Tools for Evaluating the Ecological Importance of Antimicrobial Peptides. Pharmaceuticals 2018, 11, 68. https://doi.org/10.3390/ph11030068
Ongey EL, Pflugmacher S, Neubauer P. Bioinspired Designs, Molecular Premise and Tools for Evaluating the Ecological Importance of Antimicrobial Peptides. Pharmaceuticals. 2018; 11(3):68. https://doi.org/10.3390/ph11030068
Chicago/Turabian StyleOngey, Elvis Legala, Stephan Pflugmacher, and Peter Neubauer. 2018. "Bioinspired Designs, Molecular Premise and Tools for Evaluating the Ecological Importance of Antimicrobial Peptides" Pharmaceuticals 11, no. 3: 68. https://doi.org/10.3390/ph11030068
APA StyleOngey, E. L., Pflugmacher, S., & Neubauer, P. (2018). Bioinspired Designs, Molecular Premise and Tools for Evaluating the Ecological Importance of Antimicrobial Peptides. Pharmaceuticals, 11(3), 68. https://doi.org/10.3390/ph11030068