Targeting SARS-CoV-2 Variants with Nucleic Acid Therapeutic Nanoparticle Conjugates
Abstract
:1. Introduction
2. Results
2.1. Role of RNA Structures in the Function of Coronavirus
2.2. Target Site in Variants of Concern
3. Discussion
3.1. Target Site Characterization
3.2. Confirmation of Triplex Formation
3.3. Nanoparticle-Mediated Delivery
4. Methods
4.1. GWRPD Analysis
4.2. RNA Folding
4.3. Target Site for Variants of Concern
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, Design, and Chemistry in siRNA Delivery Systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef]
- Koenig, P.A.; Das, H.; Liu, H.; Kümmerer, B.M.; Gohr, F.N.; Jenster, L.M.; Schiffelers, L.D.J.; Tesfamariam, Y.M.; Uchima, M.; Wureth, J.D.; et al. Structure-Guided Multivalent Nanobodies Block SARS-CoV-2 Infection and Suppress Mutational Escape. Science 2021, 371, eabe6230. [Google Scholar] [CrossRef]
- Kang, Y.F.; Sun, C.; Zhuang, Z.; Yuan, R.Y.; Zheng, Q.; Li, J.P.; Zhou, P.P.; Chen, X.C.; Liu, Z.; Zhang, X.; et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. Am. Chem. Soc. Nano 2021, 15, 2738–2752. [Google Scholar] [CrossRef]
- Ho, W.; Gao, M.; Li, F.; Li, Z.; Zhang, X.Q.; Xu, X. Next-Generation Vaccines: Nanoparticle-Mediated DNA and mRNA Delivery. Adv. Healthc. Mater. 2021, 10, 2001812. [Google Scholar] [CrossRef]
- Chauhan, G.; Madou, M.J.; Kalra, S.; Chopra, V.; Ghosh, D.; Martinez-Chapa, S.O. Nanotechnolgy for COVID-19: Therapeutics and Vaccine Research. Am. Chem. Soc. Nano 2020, 14, 7760–7782. [Google Scholar] [CrossRef]
- Hu, T.Y.; Frieman, M.; Wolfram, J. Insights from Nanomedicine into Chloroquine Efficacy Against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delong, R.K.; Swanson, R.; Niederwerder, M.C.; Khanal, P.; Aryal, S.; Marasini, R.; Jaberi-Douraki, M.; Shaker, H.; Mazloom, R.; Schneider, S.; et al. Zn-Based Physiometacomposite Nanoparticles: Distribution, Tolerance, Imaging, and Antiviral and Anticancer Activity. Nanomedicine 2021, 16, 1857–1872. [Google Scholar] [CrossRef]
- Delong, R.K.; Dean, J.; Glaspell, G.; Jaberi-Douraki, M.; Ghosh, K.; Davis, D.; Monteiro-Riviere, N.; Chandran, P.; Nguyen, T.; Aryal, S.; et al. Amino/Amido Conjugates Form to Nanoscale Cobalt Physiometacomposite (PMC) Materials Functionally Delivering Nucleic Acid Therapeutics to Nucleus Enhancing Anticancer Activity via Ras-Targeted Protein Interference. ACS Appl. Bio Mater. 2020, 3, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Leibowitz, J.L. The Structure and Functions of Coronavirus Genomic 3′ and 5′ Ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef]
- Miller, P.S.; Bi, G.; Kipp, S.A.; Fok, V.; DeLong, R.K. Triplex Formation by a Psoralen-Conjugated Oligodeoxyribonucleotide Containing the Base Analog 8-Oxo-Adenine. Nucleic Acids Res. 1996, 24, 730–736. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; DeLong, R.K.; Wickstrom, E.; Kligshteyn, M.; Demirdji, S.H.; Caruthers, M.H.; Juliano, R.L. Interactions between Single-Stranded DNA Binding Protein and Oligonucleotide Analogs with Different Backbone Chemistries. J. Mol. Recognit. JMR 1997, 10, 101–107. [Google Scholar] [CrossRef]
- Delong, R.K.; Miller, P.S. Inhibition of Human Collagenase Activity by Antisense Oligonucleoside Methylphosphonates. Antisense Nucleic Acid Drug Dev. 1996, 6, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kierzek, E.; Loo, Z.P.; Antonio, M.; Yau, Y.H.; Chuah, Y.W.; Geifman-Shochat, S.; Kierzek, R.; Chen, G. Recognition of RNA Duplexes by Chemically Modified Triplex-Forming Oligonucleotides. Nucleic Acids Res. 2013, 41, 6664–6673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesy, J.; Patil, K.M.; Kumar, S.R.; Shu, Z.; Yong, H.Y.; Zimmermann, L.; Ong, A.A.L.; Toh, D.F.K.; Krishna, M.S.; Yang, L.; et al. A Short Chemically Modified dsRNA-Binding PNA (dbPNA) Inhibits Influenza Viral Replication by Targeting Viral RNA Panhandle Structure. Bioconjugate Chem. 2019, 30, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P. Pervasive RNA Secondary Structure in the Genomes of SARS-CoV-2 and Other Coronaviruses. mBio 2020, 11. [Google Scholar] [CrossRef]
- Manfredonia, I.; Incarnato, D. Structure and Regulation of Coronavirus Genomes: State-of-the-art and Novel Insights from SARS-CoV-2 Studies. Biochem. Soc. Trans. 2021, 49, 341–342. [Google Scholar] [CrossRef]
- Huston, N.C.; Wan, H.; Strine, M.S.; de Cesares Araujo Tavares, R.; Wilen, C.B.; Pyle, A.M. Comprehensive in vivo Secondary Structure of the SARS-CoV-2 Genome Reveals Novel Regulatory Motifs and Mechanisms. Mol. Cell 2021, 81, 584–598.e5. [Google Scholar] [CrossRef]
- Li, L.; Kang, H.; Liu, P.; Makkinje, N.; Williamson, S.T.; Leibowitz, J.L.; Giedroc, D.P. Structural Liability in Stem-Loop 1 Drives a 5′ UTR—3′ UTR Interaction in Coronavirus Replication. J. Mol. Biol. 2008, 377, 790–803. [Google Scholar] [CrossRef]
- Madhugiri, R.; Karl, N.; Peterson, D.; Lamkiewicz, K.; Fricke, M.; Wend, U.; Scheuer, R.; Marz, M.; Ziebuhr, J. Structural and Functional Conservation of cis-acting RNA Elements in Coronavirus 5′-Terminal Genome Regions. Virology 2018, 517, 44–55. [Google Scholar] [CrossRef]
- Liu, P.; Li, L.; Millership, J.J.; Kang, H.; Leibowitz, J.L.; Giedroc, D.P. A U-Turn Motif-Containing Stem-Loop in the Coronavirus 5′ Untranslated Regions Plays a Functional Role in Replication. RNA 2007, 13, 763–780. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Liu, P.; Giedroc, D.P.; Leibowitz, J. Mouse Hepatitis Virus Stem-Loop 4 Functions as a Spacer Element Required to Drive Subgenomic RNA Synthesis. J. Virol. 2011, 85, 9199–9209. [Google Scholar] [CrossRef] [Green Version]
- Manfredonia, I.; Nithin, C.; Ponce-Salvatierra, A.; Ghosh, P.; Wirecki, T.K.; Marinus, T.; Ogando, N.S.; Snijder, E.J.; van Hemert, M.J.; Bujnicki, J.M.; et al. Genome-Wide Mapping of SARS-CoV-2 RNA Structures Identifies Therapeutically-Relevant Elements. Nucleic Acids Res. 2020, 48, 12436–12452. [Google Scholar] [CrossRef]
- Plant, E.P.; Pérez-Alvarado, G.C.; Jacobs, J.L.; Mukhopadhyay, B.; Hennig, M.; Dinman, J.D. A Three-Stemmed mRNA Pseudoknot in the SARS Coronavirus Frameshift Signal. PLoS Biol. 2005, 3, e172. [Google Scholar] [CrossRef] [Green Version]
- Kelly, J.A.; Olson, A.N.; Neupane, K.; Munshi, S.; Emeterio, J.S.; Pollack, L.; Woodside, M.T.; Dinman, J.D. Structural and Functional Conservation of the Programmed −1 Ribosomal Frameshift Signal of SARS-CoV-2. J. Biol. Chem. 2020, 29, 10741–10748. [Google Scholar] [CrossRef]
- Ziv, O.; Price, J.; Shalamova, L.; Kamenova, T.; Goodfellow, I.; Weber, F.; Miska, E.A. The Short- and Long-Range RNA-RNA Interactome of SARS-CoV-2. Mol. Cell 2020, 80, 1067–1077.e5. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Igel, H.; Baertsch, R.; Haussler, D.; Ares, M., Jr.; Scott, W.G. The Structure of a Rigorously Conserved RNA Element within the SARS Virus Genome. PLoS Biol. 2004, 3. [Google Scholar] [CrossRef]
- Hsue, B.; Masters, P.S. A Bulged Stem-Loop Structure in the 3′ Untranslated Region of the Genome of the Coronavirus Mouse Hepatitis Virus is Essential for Replication. J. Virol. 1997, 71, 7567–7578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goebel, S.J.; Taylor, J.; Masters, P.S. The 3′ cis-acting Genomic Replication Element of the Severe Acute Respiratory Syndrome Coronavirus Can Function in the Murine Coronavirus Genome. J. Virol. 2004, 78, 7846–7851. [Google Scholar] [CrossRef] [Green Version]
- Mariano, G.; Farthing, R.J.; Lale-Farjat, S.L.M.; Bergeron, J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 605236. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and Evasion of Type I Interferon Responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Lei, J.; Kusov, Y.; Hilgenfeld, R. Nsp3 of Coronaviruses: Structures and Functions of a Large Multi-Domain Protein. Antivir. Res. 2018, 149, 58–74. [Google Scholar] [CrossRef]
- Huslig, G.; Marchell, N.; Hoffman, A.; Park, S.; Choi, S.O.; DeLong, R.K. Comparing the Effects of Physiologically-Based Metal Oxide Nanoparticles on Ribonucleic Acid and RAS/RBD-Targeted Triplex and Aptamer Interactions. Biochem. Biophys. Res. Commun. 2019, 517, 43–48. [Google Scholar] [CrossRef]
- Darwish, M.A. Stability of Nucleic Acid Secondary Structures and Their Contribution to Gene Expression. Ph.D. Thesis, Seton Hall University, South Orange, NJ, USA, 2010. [Google Scholar]
- Xu, J.; Zhang, X.; Zhou, S.; Shen, J.; Yang, D.; Wu, J.; Li, X.; Li, M.; Huang, X.; Sealy, J.E.; et al. A DNA Aptamer Efficiently Inhibits the Infectivity of Bovine Herpesvirus 1 by Blocking Viral Entry. Sci. Rep. 2017, 7, 11796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parhiz, H.; Shier, W.T.; Ramezani, M. From Rationally Designed Polymeric and Peptidic Systems to Sophisticated Gene Delivery Nano-vectors. Int. J. Pharm. 2013, 457, 237–259. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.K.; Vermerris, W. Recent Advances in Nanomaterial for Gene Delivery—A Review. Nanomaterials 2017, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Chandra, S.; Nguyen, H.M.; Wiltz, K.; Hall, N.; Chaudhry, S.; Olverson, G.; Mandal, T.; Dash, S.; Kundu, A. Apatamer-Functionalized Hybrid Nanoparticles to Enhance the Delivery of Doxorubicin into Breast Cancer Cells by Silencing P-glycoprotein. J. Cancer Treat. Diagn. 2020, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.K.; Kundu, A.K.; Hazari, S.; Chandra, S.; Bao, L.; Ooms, T.; Morris, G.F.; Wu, T.; Mandal, T.K.; Dash, S. Inhibition of Hepatitis C Virus Replication by Intracellular Delivery of Multiple siRNAs by Nanosomes. Mol. Ther. 2012, 20, 1724–1736. [Google Scholar] [CrossRef] [Green Version]
- Ensinck, M.; Mottais, A.; Detry, C.; Leal, T.; Carlon, M.S. On the Corner of Models and Cure: Gene Editing in Cystic Fibrosis. Front. Pharmacol. 2021, 12, 662110. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Jensen, S.A.; Day, E.S.; Ko, C.H.; Hurley, L.A.; Luciano, J.P.; Kouri, F.M.; Merkel, T.J.; Luthi, A.J.; Patel, P.C.; Cutler, J.I.; et al. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci. Transl. Med. 2013, 5, 209ra152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P. and Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- PubMed; National Library of Medicine, National Center for Biotechnology Information: Bethesda, MD, USA, 2020. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=%28coronavirus%29+OR+%28covid%29+OR+%28sars-cov%29&sort= (accessed on 22 September 2021).
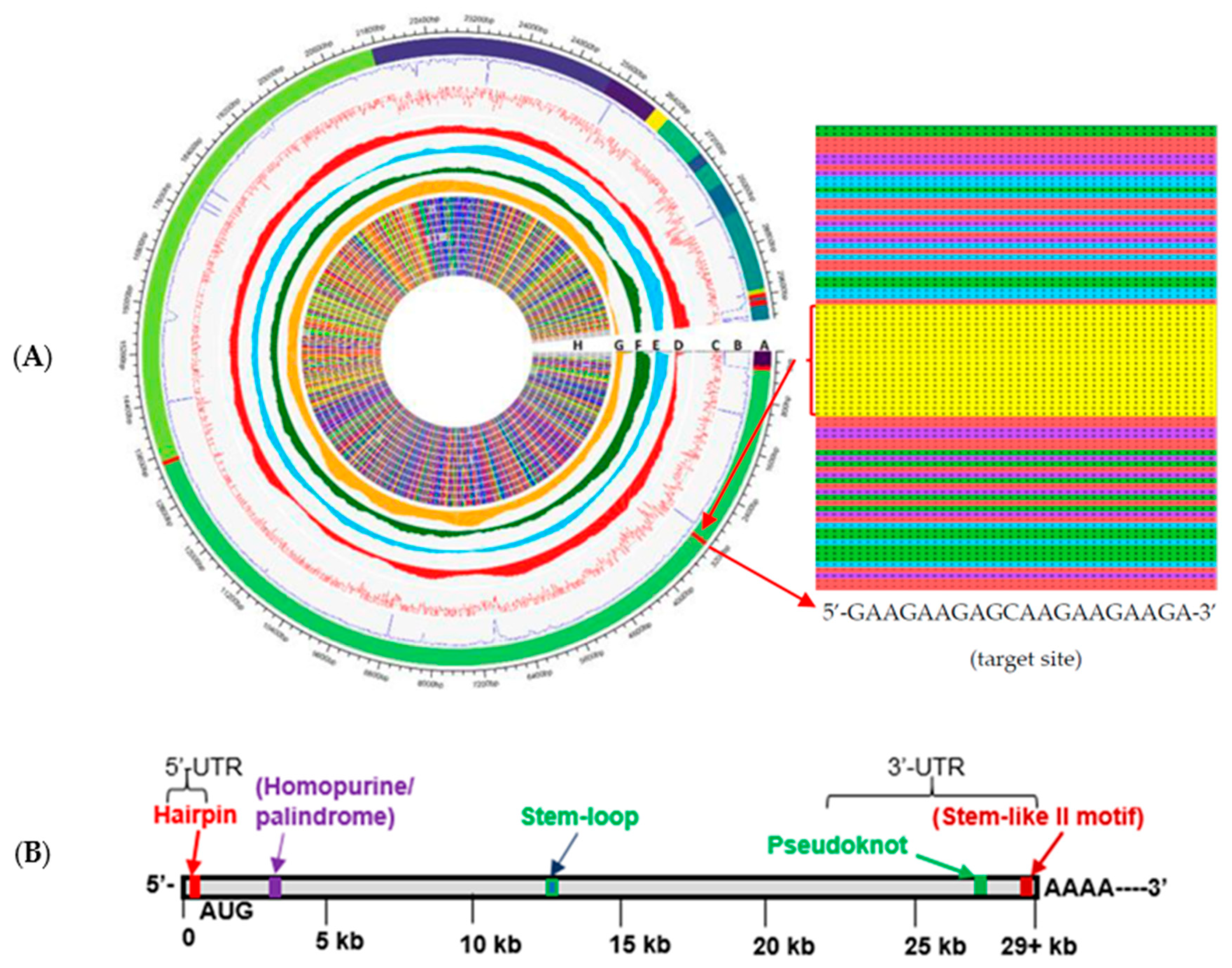

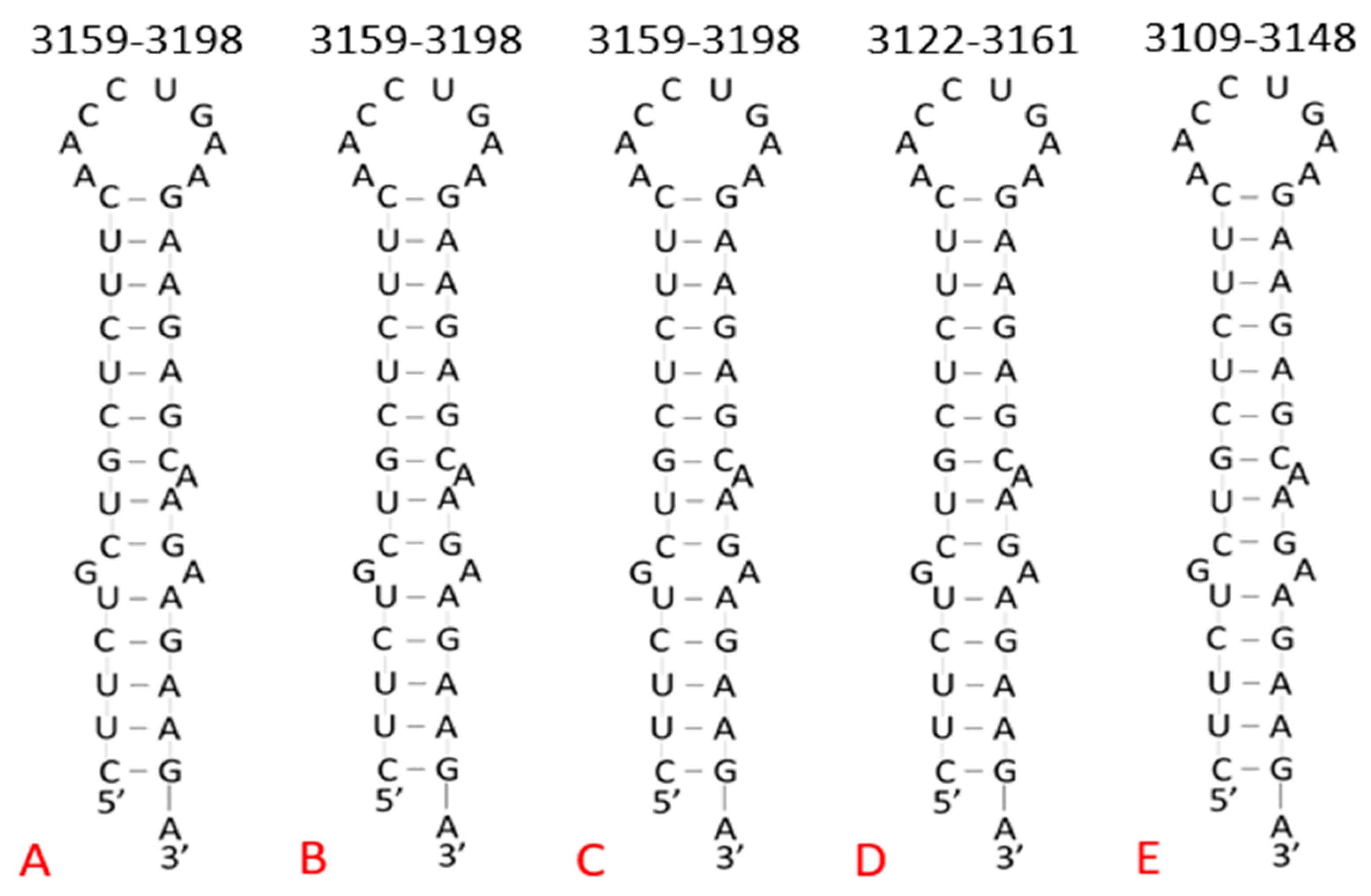
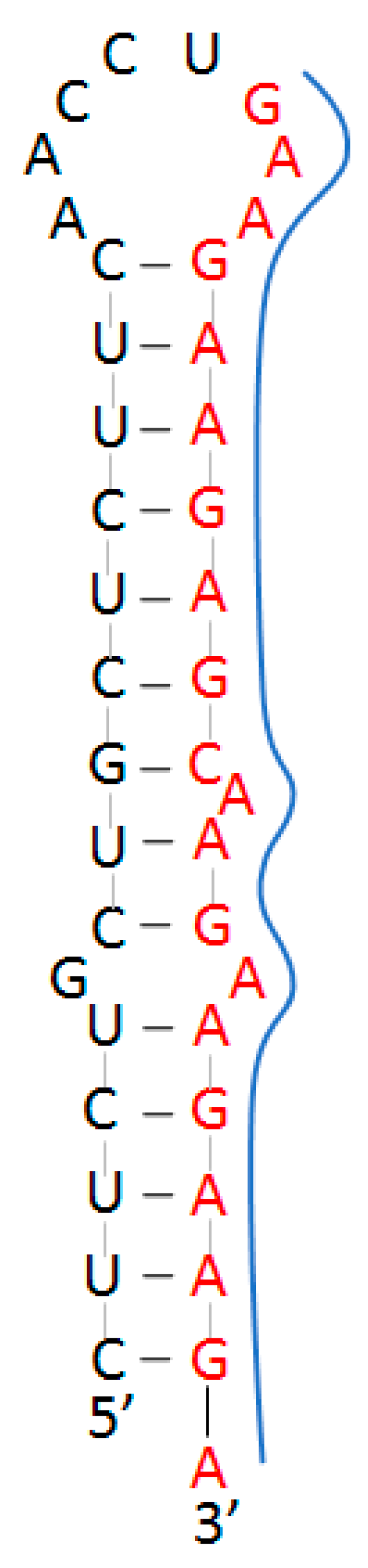
| Conserved Site (Base #) | % Conserved (Out of 1571 Variants) | Target Sequence (5′–3′) | Predicted Structure |
|---|---|---|---|
| Coding Region (3179–3198) | 99.90% | GAAGAAGAGC 1 AAGAAGAAGAAGA | Homopurine, Palindrome Stem–loop |
| 3′-UTR (29,721–29,761) | 99% | UUCACCGAGGCCACGCGGAGUACGAUCGAGUGUACAGUGA | Hairpin |
| Coding Region (13,468–13,496) | 99% | CGGUGUAAGUGCAGCCCGUCUUACACCG | Stem–loop |
| Coding Region (29,619–29,644) | 98% | GGCCCACACTGGCTTTCCATTC | Pseudoknot |
| 5′-Leader (231–265) | >97% | UCAUCAGCACAUCUAGGUUUCGUCCGGGUGUGACCGAAAGGUAA | Hairpin |
| Country and Variant (Base #) | Target Sequence 1 (5′–3′) | Sequence Length | GenBank Accession Number |
|---|---|---|---|
| Wuhan Reference (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | NC_045512.2 |
| Brazil P.1 (3171–3190) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ264787.1 |
| Brazil P.1 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ169910.1 |
| Brazil P.1 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ169911.1 |
| UK B.1.1.7 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | OU029086.1 |
| UK B.1.1.7 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | OU029131.1 |
| UK B.1.1.7 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | OU029144.1 |
| Ghana B.1.351 (3179–3198) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MW598408.1 |
| South Africa B.1.351 (3142–3161) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ376663.1 |
| Djibouti B.1.351 (3125–3144) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ520096.1 |
| India B.1.617.2 (3153–3172) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ558086.1 |
| India B.1.617.2 (3129–3148) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ340535.1 |
| India B.1.617.2 (3154–3173) | GAAGAAGAGCAAGAAGAAGAAGA | 20 nt | MZ558154.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huber, H.F.; Jaberi-Douraki, M.; DeVader, S.; Aparicio-Lopez, C.; Nava-Chavez, J.; Xu, X.; Millagaha Gedara, N.I.; Gaudreault, N.N.; Delong, R.K. Targeting SARS-CoV-2 Variants with Nucleic Acid Therapeutic Nanoparticle Conjugates. Pharmaceuticals 2021, 14, 1012. https://doi.org/10.3390/ph14101012
Huber HF, Jaberi-Douraki M, DeVader S, Aparicio-Lopez C, Nava-Chavez J, Xu X, Millagaha Gedara NI, Gaudreault NN, Delong RK. Targeting SARS-CoV-2 Variants with Nucleic Acid Therapeutic Nanoparticle Conjugates. Pharmaceuticals. 2021; 14(10):1012. https://doi.org/10.3390/ph14101012
Chicago/Turabian StyleHuber, Hanah F., Majid Jaberi-Douraki, Sarah DeVader, Cesar Aparicio-Lopez, Juliet Nava-Chavez, Xuan Xu, Nuwan Indika Millagaha Gedara, Natasha N. Gaudreault, and Robert K. Delong. 2021. "Targeting SARS-CoV-2 Variants with Nucleic Acid Therapeutic Nanoparticle Conjugates" Pharmaceuticals 14, no. 10: 1012. https://doi.org/10.3390/ph14101012
APA StyleHuber, H. F., Jaberi-Douraki, M., DeVader, S., Aparicio-Lopez, C., Nava-Chavez, J., Xu, X., Millagaha Gedara, N. I., Gaudreault, N. N., & Delong, R. K. (2021). Targeting SARS-CoV-2 Variants with Nucleic Acid Therapeutic Nanoparticle Conjugates. Pharmaceuticals, 14(10), 1012. https://doi.org/10.3390/ph14101012








