Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells
Abstract
:1. Introduction
2. Results
2.1. Gene and Protein Expression Profiles of PD-L1 and Each ERM in JEG-3 Cells
2.2. Subcellular Localization of Ezrin, Radixin, and PD-L1 in JEG-3 Cells
2.3. PD-L1 Interacts with Ezrin and Radixin in JEG-3 Cells
2.4. Effect of siRNAs against Ezrin and Radixin, Respectively, on Expression Levels of Target mRNAs and Proteins in JEG-3 Cells
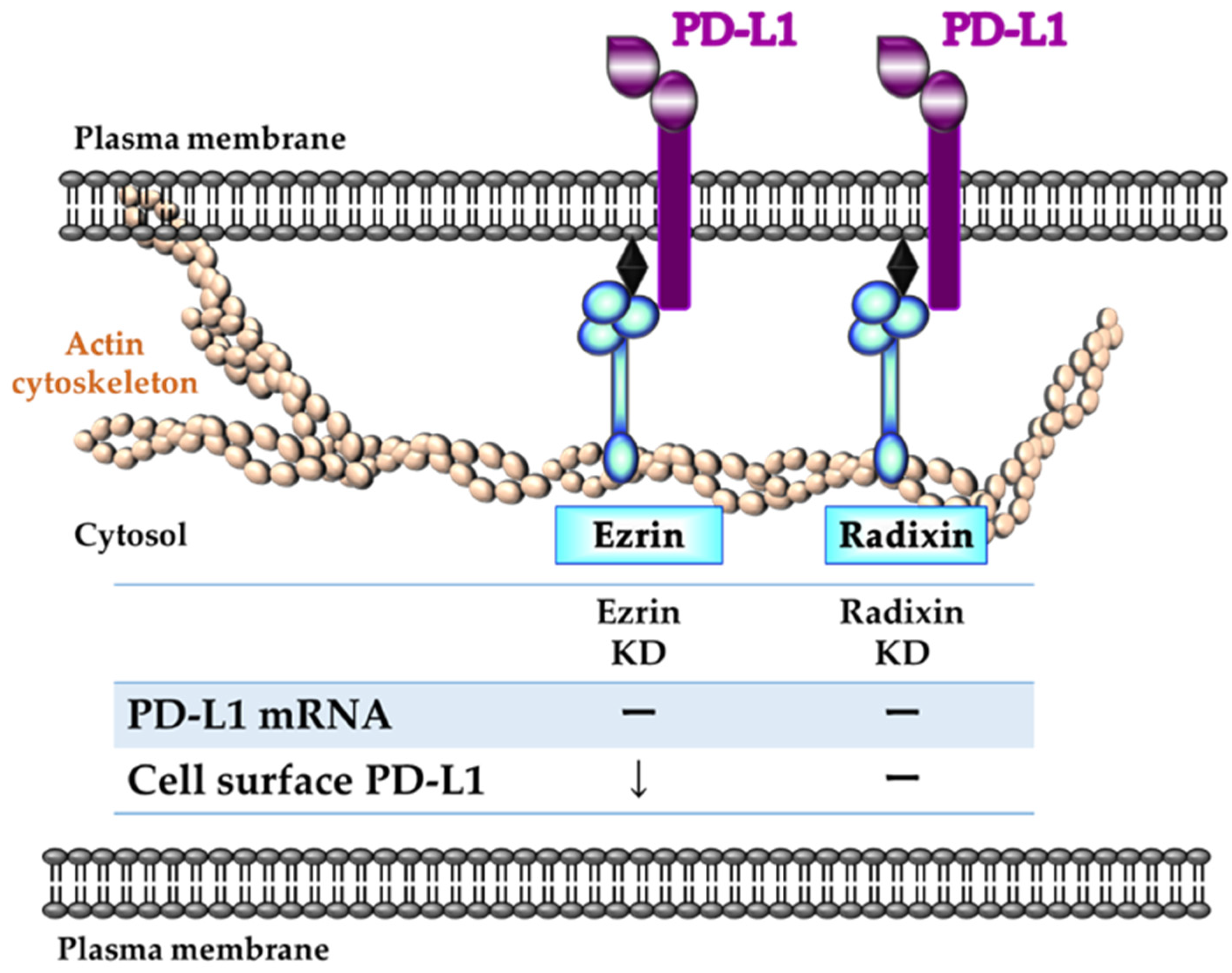
2.5. Effect of ERM Silencing on mRNA and Cell Surface Expressions of PD-L1 in JEG-3 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. siRNA Treatment
4.3. Extraction of Total RNA and Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.4. Confocal Laser Scanning Microscopy (CLSM) Analysis
4.4.1. Single Immunofluorescence Staining
4.4.2. Double Immunofluorescence Staining
4.5. Cell Viability Assay
4.6. Protein Isolation
4.7. Western Blotting
4.8. Immunoprecipitation Assay
4.9. Flow Cytometry Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldstein, D.P.; Berkowitz, R.S. Current management of gestational trophoblastic neoplasia. Hematol. Oncol. Clin. N. Am. 2012, 26, 111–131. [Google Scholar] [CrossRef]
- Seckl, M.J.; Sebire, N.J.; Berkowitz, R.S. Gestational trophoblastic disease. Lancet 2010, 376, 717–729. [Google Scholar] [CrossRef]
- Ghorani, E.; Kaur, B.; Fisher, R.A.; Short, D.; Joneborg, U.; Carlson, J.W.; Akarca, A.; Marafioti, T.; Quezada, S.A.; Sarwar, N.; et al. Pembrolizumab is effective for drug-resistant gestational trophoblastic neoplasia. Lancet 2017, 390, 2343–2345. [Google Scholar] [CrossRef] [Green Version]
- Bolze, P.A.; Attia, J.; Massardier, J.; Seckl, M.J.; Massuger, L.; van Trommel, N.; Niemann, I.; Hajri, T.; Schott, A.M.; Golfier, F.; et al. Formalised consensus of the European Organisation for Treatment of Trophoblastic Diseases on management of gestational trophoblastic diseases. Eur. J. Cancer 2015, 51, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J. Hematol. Oncol. 2013, 6, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohaegbulam, K.C.; Assal, A.; Lazar-Molnar, E.; Yao, Y.; Zang, X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 2015, 21, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, J.F.; Ceballos, P.; Lu, Z.Y. Immune precision medicine for cancer: A novel insight based on the efficiency of immune effector cells. Cancer Commun. 2019, 39, 34. [Google Scholar] [CrossRef] [Green Version]
- Veras, E.; Kurman, R.J.; Wang, T.L.; Shih, I.M. PD-L1 Expression in Human Placentas and Gestational Trophoblastic Diseases. Int. J. Gynecol. Pathol. 2017, 36, 146–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inaguma, S.; Wang, Z.; Lasota, J.; Sarlomo-Rikala, M.; McCue, P.A.; Ikeda, H.; Miettinen, M. Comprehensive Immunohistochemical Study of Programmed Cell Death Ligand 1 (PD-L1): Analysis in 5536 Cases Revealed Consistent Expression in Trophoblastic Tumors. Am. J. Surg. Pathol. 2016, 40, 1133–1142. [Google Scholar] [CrossRef] [Green Version]
- Bolze, P.A.; Patrier, S.; Massardier, J.; Hajri, T.; Abbas, F.; Schott, A.M.; Allias, F.; Devouassoux-Shisheboran, M.; Freyer, G.; Golfier, F.; et al. PD-L1 Expression in Premalignant and Malignant Trophoblasts from Gestational Trophoblastic Diseases Is Ubiquitous and Independent of Clinical Outcomes. Int. J. Gynecol. Cancer 2017, 27, 554–561. [Google Scholar] [CrossRef]
- Zong, L.; Zhang, M.; Wang, W.; Wan, X.; Yang, J.; Xiang, Y. PD-L1, B7-H3 and VISTA are highly expressed in gestational trophoblastic neoplasia. Histopathology 2019, 75, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Nixon, N.A.; Blais, N.; Ernst, S.; Kollmannsberger, C.; Bebb, G.; Butler, M.; Smylie, M.; Verma, S. Current landscape of immunotherapy in the treatment of solid tumours, with future opportunities and challenges. Curr. Oncol. 2018, 25, e373–e384. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Johnson, B.A., 3rd; Lutz, E.R.; Laheru, D.A.; Jaffee, E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 2017, 17, 209–222. [Google Scholar] [CrossRef]
- Qin, W.; Hu, L.; Zhang, X.; Jiang, S.; Li, J.; Zhang, Z.; Wang, X. The Diverse Function of PD-1/PD-L Pathway beyond Cancer. Front. Immunol. 2019, 10, 2298. [Google Scholar] [CrossRef] [PubMed]
- You, B.; Bolze, P.A.; Lotz, J.P.; Massardier, J.; Gladieff, L.; Joly, F.; Hajri, T.; Maucort-Boulch, D.; Bin, S.; Rousset, P.; et al. Avelumab in Patients with Gestational Trophoblastic Tumors with Resistance to Single-Agent Chemotherapy: Cohort A of the TROPHIMMUN Phase II Trial. J. Clin. Oncol. 2020, 38, 3129–3137. [Google Scholar] [CrossRef]
- Choi, M.C.; Oh, J.; Lee, C. Effective anti-programmed cell death 1 treatment for chemoresistant gestational trophoblastic neoplasia. Eur. J. Cancer 2019, 121, 94–97. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, S.; Akbay, E.A.; Li, Y.Y.; Herter-Sprie, G.S.; Buczkowski, K.A.; Richards, W.G.; Gandhi, L.; Redig, A.J.; Rodig, S.J.; Asahina, H.; et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 2016, 7, 10501. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, X. Study and analysis of antitumor resistance mechanism of PD1/PD-L1 immune checkpoint blocker. Cancer Med. 2020, 9, 8086–8121. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ruiz, E.; Melero, I.; Kopecka, J.; Sarmento-Ribeiro, A.B.; Garcia-Aranda, M.; De Las Rivas, J. Cancer immunotherapy resistance based on immune checkpoints inhibitors: Targets, biomarkers, and remedies. Drug Resist. Updates 2020, 53, 100718. [Google Scholar] [CrossRef]
- Wang, Y.N.; Lee, H.H.; Hsu, J.L.; Yu, D.; Hung, M.C. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J. Biomed. Sci. 2020, 27, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, S.M.; Zhou, J.; Wang, Y.; Nie, R.C.; Chen, J.W.; Xie, D. Recent Findings in the Posttranslational Modifications of PD-L1. J. Oncol. 2020, 2020, 5497015. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef] [Green Version]
- Ogihara, T.; Mizoi, K.; Kamioka, H.; Yano, K. Physiological Roles of ERM Proteins and Transcriptional Regulators in Supporting Membrane Expression of Efflux Transporters as Factors of Drug Resistance in Cancer. Cancers 2020, 12, 3352. [Google Scholar] [CrossRef]
- Kobori, T.; Harada, S.; Nakamoto, K.; Tokuyama, S. Mechanisms of P-glycoprotein alteration during anticancer treatment: Role in the pharmacokinetic and pharmacological effects of various substrate drugs. J. Pharmacol. Sci. 2014, 125, 242–254. [Google Scholar] [CrossRef] [Green Version]
- Luciani, F.; Molinari, A.; Lozupone, F.; Calcabrini, A.; Lugini, L.; Stringaro, A.; Puddu, P.; Arancia, G.; Cianfriglia, M.; Fais, S. P-glycoprotein-actin association through ERM family proteins: A role in P-glycoprotein function in human cells of lymphoid origin. Blood 2002, 99, 641–648. [Google Scholar] [CrossRef]
- Asp, N.; Kvalvaag, A.; Sandvig, K.; Pust, S. Regulation of ErbB2 localization and function in breast cancer cells by ERM proteins. Oncotarget 2016, 7, 25443–25460. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Su, Y.; Xu, B. Rho-associated protein kinase-dependent moesin phosphorylation is required for PD-L1 stabilization in breast cancer. Mol. Oncol. 2020, 14, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kwon, O.K.; Han, M.S.; Lee, Y.M.; Kim, S.W.; Kim, K.M.; Lee, T.; Lee, S.; Bae, J.S. Role of moesin in HMGB1-stimulated severe inflammatory responses. Thromb. Haemost. 2015, 114, 350–363. [Google Scholar] [PubMed]
- Wang, Q.; Fan, A.; Yuan, Y.; Chen, L.; Guo, X.; Huang, X.; Huang, Q. Role of Moesin in Advanced Glycation End Products-Induced Angiogenesis of Human Umbilical Vein Endothelial Cells. Sci. Rep. 2016, 6, 22749. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, E.R.; Bonfim-Melo, A.; Cordero, E.M.; Mortara, R.A. ERM Proteins Play Distinct Roles in Cell Invasion by Extracellular Amastigotes of Trypanosoma cruzi. Front. Microbiol. 2017, 8, 2230. [Google Scholar] [CrossRef] [PubMed]
- Zaman, R.; Lombardo, A.; Sauvanet, C.; Viswanatha, R.; Awad, V.; Bonomo, L.E.; McDermitt, D.; Bretscher, A. Effector-mediated ERM activation locally inhibits RhoA activity to shape the apical cell domain. J. Cell Biol. 2021, 220, e202007146. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef] [Green Version]
- Viswanatha, R.; Wayt, J.; Ohouo, P.Y.; Smolka, M.B.; Bretscher, A. Interactome analysis reveals ezrin can adopt multiple conformational states. J. Biol. Chem. 2013, 288, 35437–35451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, K.; Iizasa, H.; Sai, Y.; Horieya, S.; Lee, K.E.; Wada, M.; Deguchi, M.; Nishimura, T.; Wakayama, T.; Tamura, A.; et al. Differential expression of ezrin and CLP36 in the two layers of syncytiotrophoblast in rats. Biol. Pharm. Bull. 2010, 33, 1400–1406. [Google Scholar] [CrossRef] [Green Version]
- Berggren, S.; Gall, C.; Wollnitz, N.; Ekelund, M.; Karlbom, U.; Hoogstraate, J.; Schrenk, D.; Lennernas, H. Gene and protein expression of P-glycoprotein, MRP1, MRP2, and CYP3A4 in the small and large human intestine. Mol. Pharm. 2007, 4, 252–257. [Google Scholar] [CrossRef]
- Gerlach, J.H. Structure and function of P-glycoprotein. Cancer Treat. Res. 1989, 48, 37–53. [Google Scholar] [PubMed]
- Ogihara, T.; Kamiya, M.; Ozawa, M.; Fujita, T.; Yamamoto, A.; Yamashita, S.; Ohnishi, S.; Isomura, Y. What kinds of substrates show P-glycoprotein-dependent intestinal absorption? Comparison of verapamil with vinblastine. Drug Metab. Pharmacokinet. 2006, 21, 238–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudnicka, D.; Oszmiana, A.; Finch, D.K.; Strickland, I.; Schofield, D.J.; Lowe, D.C.; Sleeman, M.A.; Davis, D.M. Rituximab causes a polarization of B cells that augments its therapeutic function in NK-cell-mediated antibody-dependent cellular cytotoxicity. Blood 2013, 121, 4694–4702. [Google Scholar] [CrossRef]
- Ghosh, S.; Di Bartolo, V.; Tubul, L.; Shimoni, E.; Kartvelishvily, E.; Dadosh, T.; Feigelson, S.W.; Alon, R.; Alcover, A.; Haran, G. ERM-Dependent Assembly of T Cell Receptor Signaling and Co-stimulatory Molecules on Microvilli prior to Activation. Cell Rep. 2020, 30, 3434–3447.e3436. [Google Scholar] [CrossRef] [Green Version]
- Hoshi, Y.; Uchida, Y.; Kuroda, T.; Tachikawa, M.; Couraud, P.O.; Suzuki, T.; Terasaki, T. Distinct roles of ezrin, radixin and moesin in maintaining the plasma membrane localizations and functions of human blood-brain barrier transporters. J. Cereb. Blood Flow Metab. 2020, 40, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, J.; Zhu, X.; Wang, W.; Yang, Q. The effect of sphingomyelin synthase 2 (SMS2) deficiency on the expression of drug transporters in mouse brain. Biochem. Pharmacol. 2011, 82, 287–294. [Google Scholar] [CrossRef]
- Kobori, T.; Fujiwara, S.; Miyagi, K.; Harada, S.; Nakamoto, K.; Nakagawa, T.; Takahashi, H.; Narita, M.; Tokuyama, S. Involvement of moesin in the development of morphine analgesic tolerance through P-glycoprotein at the blood-brain barrier. Drug Metab. Pharmacokinet. 2014, 29, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Otsuka, K.; Kato, Y.; Kawabata, H.; Ohmori, S.; Arakawa, H.; Ogihara, T. Different regulation of P-glycoprotein function between Caco-2 and Caki-1 cells by ezrin, radixin and moesin proteins. J. Pharm. Pharmacol. 2016, 68, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Kobori, T.; Tameishi, M.; Tanaka, C.; Urashima, Y.; Obata, T. Subcellular distribution of ezrin/radixin/moesin and their roles in the cell surface localization and transport function of P-glycoprotein in human colon adenocarcinoma LS180 cells. PLoS ONE 2021, 16, e0250889. [Google Scholar] [CrossRef]
- Kobori, T.; Hamasaki, S.; Kitaura, A.; Yamazaki, Y.; Nishinaka, T.; Niwa, A.; Nakao, S.; Wake, H.; Mori, S.; Yoshino, T.; et al. Interleukin-18 Amplifies Macrophage Polarization and Morphological Alteration, Leading to Excessive Angiogenesis. Front. Immunol. 2018, 9, 334. [Google Scholar] [CrossRef]
- Hamasaki, S.; Kobori, T.; Yamazaki, Y.; Kitaura, A.; Niwa, A.; Nishinaka, T.; Nishibori, M.; Mori, S.; Nakao, S.; Takahashi, H. Effects of scavenger receptors-1 class A stimulation on macrophage morphology and highly modified advanced glycation end product-protein phagocytosis. Sci. Rep. 2018, 8, 5901. [Google Scholar] [CrossRef]
- Lall, N.; Henley-Smith, C.J.; De Canha, M.N.; Oosthuizen, C.B.; Berrington, D. Viability Reagent, PrestoBlue, in Comparison with Other Available Reagents, Utilized in Cytotoxicity and Antimicrobial Assays. Int. J. Microbiol. 2013, 2013, 420601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockert, J.C.; Horobin, R.W.; Colombo, L.L.; Blazquez-Castro, A. Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018, 120, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Boncler, M.; Rozalski, M.; Krajewska, U.; Podsedek, A.; Watala, C. Comparison of PrestoBlue and MTT assays of cellular viability in the assessment of anti-proliferative effects of plant extracts on human endothelial cells. J. Pharmacol. Toxicol. Methods 2014, 69, 9–16. [Google Scholar] [CrossRef]
- Xu, M.; McCanna, D.J.; Sivak, J.G. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J. Pharmacol. Toxicol. Methods 2015, 71, 1–7. [Google Scholar] [CrossRef]
- Kobori, T.; Harada, S.; Nakamoto, K.; Tokuyama, S. Changes in PtdIns(4,5)P2 induced by etoposide treatment modulates small intestinal P-glycoprotein via radixin. Biol. Pharm. Bull. 2014, 37, 1124–1131. [Google Scholar] [CrossRef] [Green Version]
- Kobori, T.; Harada, S.; Nakamoto, K.; Tokuyama, S. Radixin influences the changes in the small intestinal p-glycoprotein by Etoposide treatment. Biol. Pharm. Bull. 2013, 36, 1822–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene | Primer Sequence (5′→3′) |
|---|---|
| h-β-actin (forward) | TGGCACCCAGCACAATGAA |
| h-β-actin (reverse) | CTAAGTCATAGTCCGCCTAGAAGCA |
| h-ezrin (forward) | ACCATGGATGCAGAGCTGGAG |
| h-ezrin (reverse) | CATAGTGGAGGCCAAAGTACCACA |
| h-radixin (forward) | GAATTTGCCATTCAGCCCAATA |
| h-radixin (reverse) | GCCATGTAGAATAACCTTTGCTGTC |
| h-moesin (forward) | CCGAATCCAAGCCGTGTGTA |
| h-moesin (reverse) | GGCAAACTCCAGCTCTGCATC |
| h-PD-L1 (forward) | CAATGTGACCAGCACACTGAGAA |
| h-PD-L1 (reverse) | GGCATAATAAGATGGCTCCCAGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tameishi, M.; Kobori, T.; Tanaka, C.; Urashima, Y.; Ito, T.; Obata, T. Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells. Pharmaceuticals 2021, 14, 963. https://doi.org/10.3390/ph14100963
Tameishi M, Kobori T, Tanaka C, Urashima Y, Ito T, Obata T. Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells. Pharmaceuticals. 2021; 14(10):963. https://doi.org/10.3390/ph14100963
Chicago/Turabian StyleTameishi, Mayuka, Takuro Kobori, Chihiro Tanaka, Yoko Urashima, Takuya Ito, and Tokio Obata. 2021. "Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells" Pharmaceuticals 14, no. 10: 963. https://doi.org/10.3390/ph14100963
APA StyleTameishi, M., Kobori, T., Tanaka, C., Urashima, Y., Ito, T., & Obata, T. (2021). Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells. Pharmaceuticals, 14(10), 963. https://doi.org/10.3390/ph14100963







