HDAC4 Is Indispensable for Reduced Slow Myosin Expression at the Early Stage of Hindlimb Unloading in Rat Soleus Muscle
Abstract
:1. Introduction
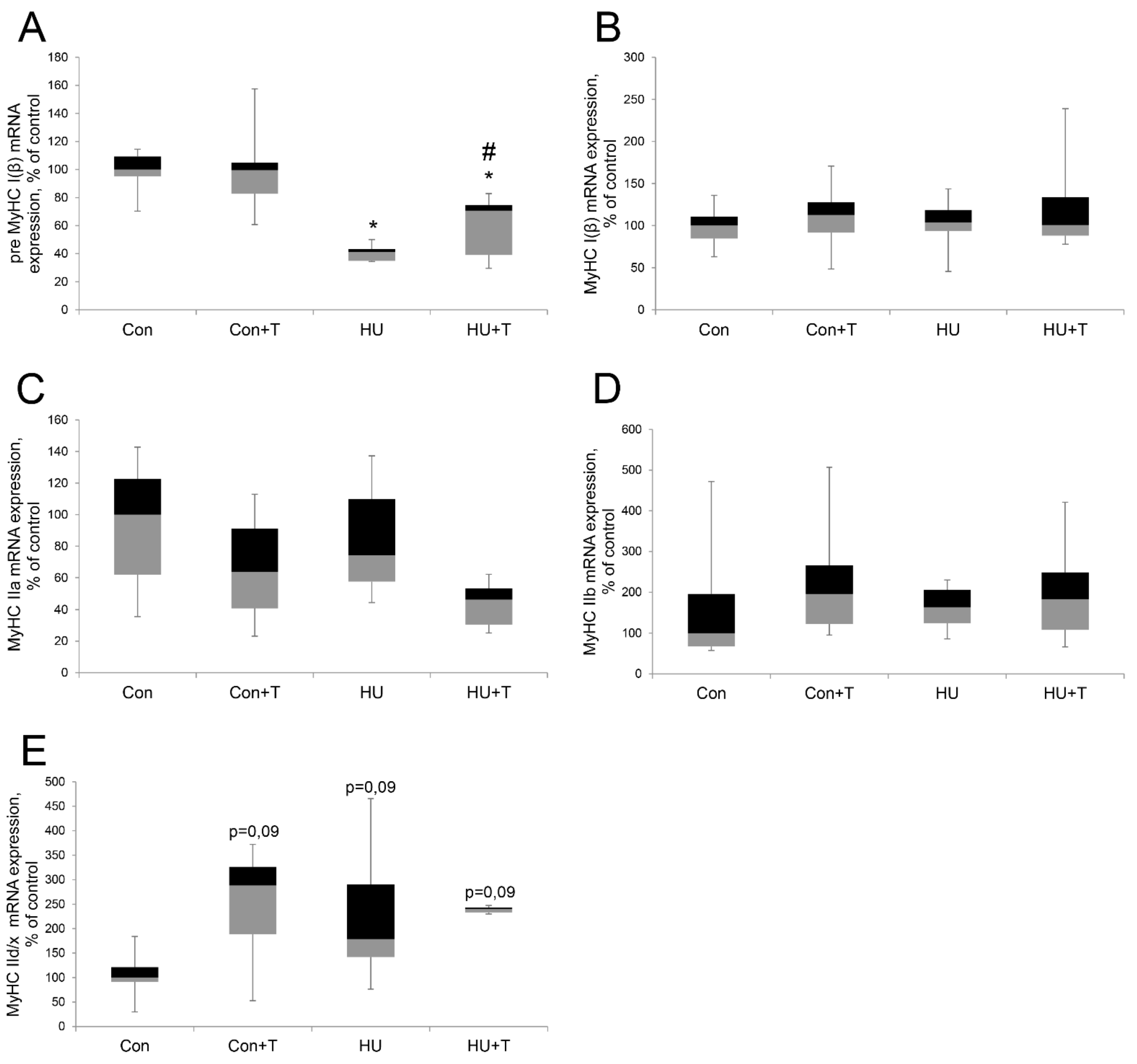
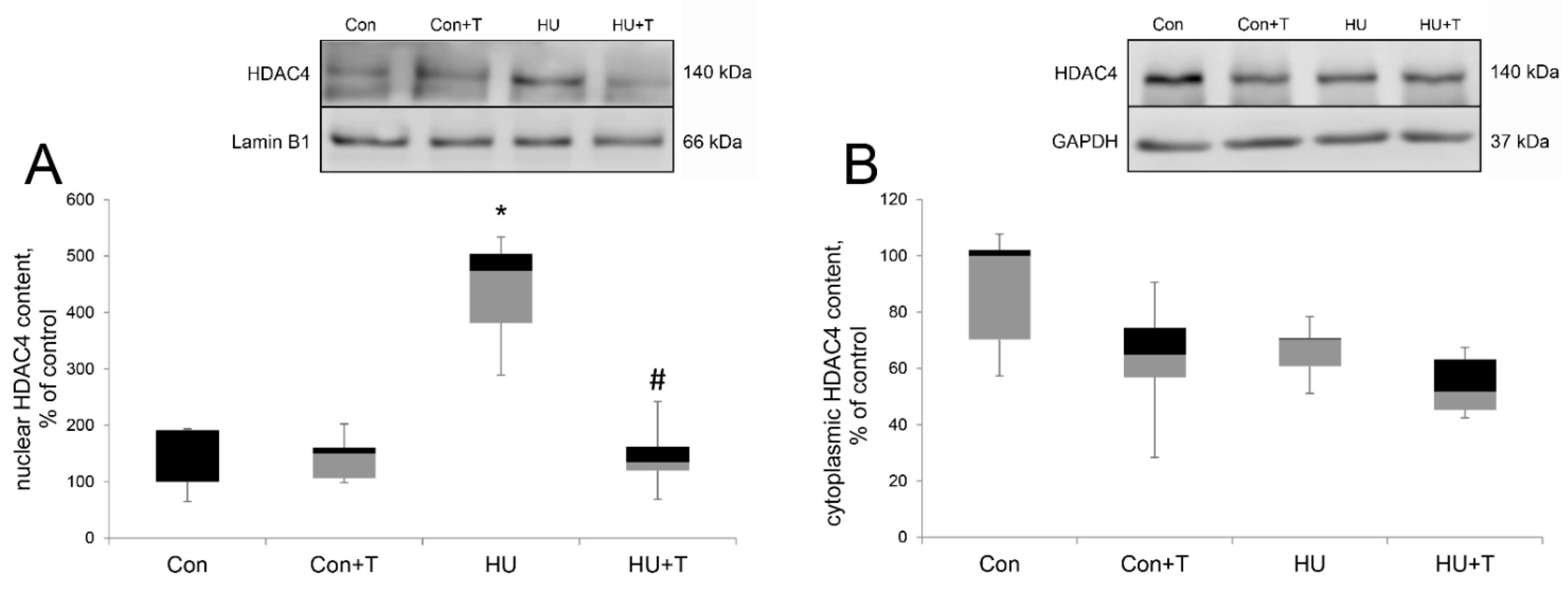
2. Results
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Study Design
4.3. Hindlimb Suspension Protocol
4.4. Protein Extraction and Western Blot Analysis
4.5. Co-Immunoprecipitation
4.6. RNA Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pette, D.; Staron, R.S. Transitions of muscle fiber phenotypic profiles. Histochem. Cell Biol. 2001, 115, 359–372. [Google Scholar] [CrossRef]
- Stevens, L.; Sultan, K.R.; Peuker, H.; Gohlsch, B.; Mounier, Y.; Pette, D. Time-dependent changes in myosin heavy chain mRNA and protein isoforms in unloaded soleus muscle of rat. Am. J. Physiol. 1999, 277, C1044–C1049. [Google Scholar] [CrossRef] [PubMed]
- Desplanches, D.; Mayet, M.H.; Ilyina-Kakueva, E.I.; Frutoso, J.; Flandrois, R. Structural and metabolic properties of rat muscle exposed to weightlessness aboard Cosmos 1887. Eur. J. Appl. Physiol. Occup. Physiol. 1991, 63, 288–292. [Google Scholar] [CrossRef]
- Desplanches, D.; Mayet, M.H.; Sempore, B.; Flandrois, R. Structural and functional responses to prolonged hindlimb suspension in rat muscle. J. Appl. Physiol. 1987, 63, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Vilchinskaya, N.A.; Mochalova, E.P.; Nemirovskaya, T.L.; Mirzoev, T.M.; Turtikova, O.V.; Shenkman, B.S. Rapid decline in MyHC I(beta) mRNA expression in rat soleus during hindlimb unloading is associated with AMPK dephosphorylation. J. Physiol. 2017, 595, 7123–7134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desplanches, D.; Mayet, M.H.; Ilyina-Kakueva, E.I.; Sempore, B.; Flandrois, R. Skeletal muscle adaptation in rats flown on Cosmos 1667. J. Appl. Physiol. 1990, 68, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, P.; Trappe, S.; Harber, M.; Creer, A.; Mazzetti, S.; Trappe, T.; Alkner, B.; Tesch, P. Effects of 84-days of bedrest and resistance training on single muscle fibre myosin heavy chain distribution in human vastus lateralis and soleus muscles. Acta Physiol. Scand. 2005, 185, 61–69. [Google Scholar] [CrossRef]
- Giger, J.M.; Bodell, P.W.; Zeng, M.; Baldwin, K.M.; Haddad, F. Rapid muscle atrophy response to unloading: Pretranslational processes involving MHC and actin. J. Appl. Physiol. 2009, 107, 1204–1212. [Google Scholar] [CrossRef] [Green Version]
- Meissner, J.D.; Umeda, P.K.; Chang, K.C.; Gros, G.; Scheibe, R.J. Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J. Cell. Physiol. 2007, 211, 138–148. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Wu, H.; Arnold, M.A.; Shelton, J.M.; Backs, J.; McAnally, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Investig. 2007, 117, 2459–2467. [Google Scholar] [CrossRef] [Green Version]
- Sharlo, K.; Paramonova, I.; Turtikova, O.; Tyganov, S.; Shenkman, B. Plantar mechanical stimulation prevents calcineurin-NFATc1 inactivation and slow-to-fast fiber type shift in rat soleus muscle under hindlimb unloading. J. Appl. Physiol. 2019, 126, 1769–1781. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Zhang, C.L.; Lu, J.; Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000, 408, 106–111. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Zhang, C.L.; Olson, E.N. MEF2: A calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 2002, 27, 40–47. [Google Scholar] [CrossRef]
- Miska, E.A.; Karlsson, C.; Langley, E.; Nielsen, S.J.; Pines, J.; Kouzarides, T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999, 18, 5099–5107. [Google Scholar] [CrossRef]
- Sparrow, D.B.; Miska, E.A.; Langley, E.; Reynaud-Deonauth, S.; Kotecha, S.; Towers, N.; Spohr, G.; Kouzarides, T.; Mohun, T.J. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999, 18, 5085–5098. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.H.; Bertos, N.R.; Vezmar, M.; Pelletier, N.; Crosato, M.; Heng, H.H.; Th’ng, J.; Han, J.; Yang, X.J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 1999, 19, 7816–7827. [Google Scholar] [CrossRef] [Green Version]
- Lemercier, C.; Verdel, A.; Galloo, B.; Curtet, S.; Brocard, M.P.; Khochbin, S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 2000, 275, 15594–15599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 2000, 6, 233–244. [Google Scholar] [CrossRef]
- Dressel, U.; Bailey, P.J.; Wang, S.C.; Downes, M.; Evans, R.M.; Muscat, G.E. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 2001, 276, 17007–17013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, I.; Ciciliot, S.; Dyar, K.A.; Abraham, R.; Murgia, M.; Agatea, L.; Akimoto, T.; Bicciato, S.; Forcato, M.; Pierre, P.; et al. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat. Commun. 2016, 7, 12397. [Google Scholar] [CrossRef]
- Schiaffino, S.; Dyar, K.A.; Calabria, E. Skeletal muscle mass is controlled by the MRF4-MEF2 axis. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 164–167. [Google Scholar] [CrossRef]
- Pandorf, C.E.; Haddad, F.; Wright, C.; Bodell, P.W.; Baldwin, K.M. Differential epigenetic modifications of histones at the myosin heavy chain genes in fast and slow skeletal muscle fibers and in response to muscle unloading. Am. J. Physiol. Cell Physiol. 2009, 297, C6-C16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001, 11, 497–504. [Google Scholar] [CrossRef]
- Yang, H.; Menconi, M.J.; Wei, W.; Petkova, V.; Hasselgren, P.O. Dexamethasone upregulates the expression of the nuclear cofactor p300 and its interaction with C/EBPbeta in cultured myotubes. J. Cell Biochem. 2005, 94, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wei, W.; Menconi, M.; Hasselgren, P.O. Dexamethasone-induced protein degradation in cultured myotubes is p300/HAT dependent. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R337-R334. [Google Scholar] [CrossRef]
- Tobimatsu, K.; Noguchi, T.; Hosooka, T.; Sakai, M.; Inagaki, K.; Matsuki, Y.; Hiramatsu, R.; Kasuga, M. Overexpression of the transcriptional coregulator Cited2 protects against glucocorticoid-induced atrophy of C2C12 myotubes. Biochem. Biophys. Res. Commun. 2009, 378, 399–403. [Google Scholar] [CrossRef]
- Alamdari, N.; Smith, I.J.; Aversa, Z.; Hasselgren, P.O. Sepsis and glucocorticoids upregulate p300 and downregulate HDAC6 expression and activity in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R509–R520. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.F.; Greene, W.C. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J. Mol. Med. 2003, 81, 549–557. [Google Scholar] [CrossRef]
- Perrot, V.; Rechler, M.M. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol. Endocrinol. 2005, 19, 2283–2298. [Google Scholar] [CrossRef]
- Cesena, T.I.; Cardinaux, J.R.; Kwok, R.; Schwartz, J. CCAAT/enhancer-binding protein (C/EBP) beta is acetylated at multiple lysines: Acetylation of C/EBPbeta at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 2007, 282, 956–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerhart-Hines, Z.; Rodgers, J.T.; Bare, O.; Lerin, C.; Kim, S.H.; Mostoslavsky, R.; Alt, F.W.; Wu, Z.; Puigserver, P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007, 26, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wellen, K.E.; Rabinowitz, J.D. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 2016, 30, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, T.J.; Choi, M.C.; Kapur, M.; Lira, V.A.; Yan, Z.; Yao, T.P. HDAC4 regulates muscle fiber type-specific gene expression programs. Mol. Cells 2015, 38, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Randall, W.R.; Schneider, M.F. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell. Biol. 2005, 168, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Shen, T.; Randall, W.R.; Schneider, M.F. Signaling pathways in activity-dependent fiber type plasticity in adult skeletal muscle. J. Muscle Res. Cell Motil. 2005, 26, 13–21. [Google Scholar] [CrossRef]
- Rockl, K.S.; Hirshman, M.F.; Brandauer, J.; Fujii, N.; Witters, L.A.; Goodyear, L.J. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes 2007, 56, 2062–2069. [Google Scholar] [CrossRef] [Green Version]
- McGee, S.L.; Hargreaves, M. AMPK-mediated regulation of transcription in skeletal muscle. Clin. Sci. (Lond.) 2010, 118, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Fielitz, J.; McAnally, J.; Shelton, J.M.; Lemon, D.D.; McKinsey, T.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol. Cell. Biol. 2008, 28, 3600–3609. [Google Scholar] [CrossRef] [Green Version]
- Mochalova, E.P.; Belova, S.P.; Kostrominova, T.Y.; Shenkman, B.S.; Nemirovskaya, T.L. Differences in the Role of HDACs 4 and 5 in the Modulation of Processes Regulating MAFbx and MuRF1 Expression during Muscle Unloading. Int. J. Mol. Sci. 2020, 21, 4815. [Google Scholar] [CrossRef]
- Deronic, A.; Tahvili, S.; Leanderson, T.; Ivars, F. The anti-tumor effect of the quinoline-3-carboxamide tasquinimod: Blockade of recruitment of CD11b(+) Ly6C(hi) cells to tumor tissue reduces tumor growth. BMC Cancer 2016, 16, 440. [Google Scholar] [CrossRef] [Green Version]
- Olsson, A.; Bjork, A.; Vallon-Christersson, J.; Isaacs, J.T.; Leanderson, T. Tasquinimod (ABR-215050), a quinoline-3-carboxamide anti-angiogenic agent, modulates the expression of thrombospondin-1 in human prostate tumors. Mol. Cancer 2010, 9, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isaacs, J.T.; Antony, L.; Dalrymple, S.L.; Brennen, W.N.; Gerber, S.; Hammers, H.; Wissing, M.; Kachhap, S.; Luo, J.; Xing, L.; et al. Tasquinimod Is an Allosteric Modulator of HDAC4 survival signaling within the compromised cancer microenvironment. Cancer Res. 2013, 73, 1386–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belova, S.; Mochalova, E.; Nemirovskaya, T. The Role of Class IIa HDACs in the Expression of E3 Ligases ATROGIN-1/MAFbx and MuRF1 under Muscle Unloading. Biochem. (Moscow) Suppl. Ser. A Membr. Cell Biol. 2020, 14, 74–80. [Google Scholar] [CrossRef]
- Yoshihara, T.; Machida, S.; Kurosaka, Y.; Kakigi, R.; Sugiura, T.; Naito, H. Immobilization induces nuclear accumulation of HDAC4 in rat skeletal muscle. J. Physiol. Sci. 2016, 66, 337–343. [Google Scholar] [CrossRef]
- Mochalova, E.P.; Belova, S.P.; Mirzoev, T.M.; Shenkman, B.S.; Nemirovskaya, T.L. Atrogin-1/MAFbx mRNA expression is regulated by histone deacetylase 1 in rat soleus muscle under hindlimb unloading. Sci. Rep. 2019, 9, 10263. [Google Scholar] [CrossRef]
- Paramonova, I.I.; Sharlo, K.A.; Vilchinskaya, N.A.; Shenkman, B.S. The time course of muscle nuclear content of transcription factors regulating the MyHC I(β) expression in the rat soleus muscle under gravitational unloading. Biochem. (Moscow) Suppl. Ser. A Membr. Cell Biol. 2020, 14, 242–248. [Google Scholar] [CrossRef]
- Meissner, J.D.; Freund, R.; Krone, D.; Umeda, P.K.; Chang, K.C.; Gros, G.; Scheibe, R.J. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of p300 enhances myosin heavy chain I/beta gene expression via acetylation of nuclear factor of activated T cells c1. Nucleic Acids Res. 2011, 39, 5907–5925. [Google Scholar] [CrossRef] [Green Version]
- Morey-Holton, E.R.; Globus, R.K. Hindlimb unloading rodent model: Technical aspects. J. Appl. Physiol. 2002, 92, 1367–1377. [Google Scholar] [CrossRef]
- Novikov, V.E.; Ilyin, E.A. Age-related reactions of rat bones to their unloading. Aviat. Space Environ. Med. 1981, 52, 551–553. [Google Scholar]
- Belova, S.P.; Shenkman, B.S.; Kostrominova, T.Y.; Nemirovskaya, T.L. Paradoxical effect of IKKbeta inhibition on the expression of E3 ubiquitin ligases and unloading-induced skeletal muscle atrophy. Physiol. Rep. 2017, 5, e13291. [Google Scholar] [CrossRef]
- Lomonosova, Y.N.; Shenkman, B.S.; Nemirovskaya, T.L. Attenuation of unloading-induced rat soleus atrophy with the heat-shock protein inducer 17-(allylamino)-17-demethoxygeldanamycin. FASEB J. 2012, 26, 4295–4301. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene Description | Forward Primer Reverse Primer | GenBank |
|---|---|---|
| pre myh7 (MyHC I(β)) | 5′-ACTTAGCAGGCAAATCTCAGTAGC-3′ 5′-CTCGCGTTATGTTTCTCATCCGAAT-3′ | NM_017240.2 |
| Myh7 (MyHC I(β)) | 5′-ACAGAGGAAGACAGGAAGAACCTAC-3′ 5′-GGGCTTCACAGGCATCCTTAG-3′ | NM_017240.2 |
| RPL19 | 5’-GTACCCTTCCTCTTCCCTATGC-3’ 5’-CAATGCCAACTCTCGTCAACAG-3’ | NM_031103.1 |
| Myh2 (MyHC IIa) | 5′-TATCCTCAGGCTTCAAGATTTG-3′ 5′-TAAATAGAATCACATGGGGACA-3′ | NM_001135157.1 |
| Myh4 (MyHC IIb) | 5′-CTGAGGAACAATCCAACGTC-3′ 5′-TTGTGTGATTTCTTCTGTCACCT-3′ | NM_019325.1 |
| Myh1 (MyHC IId/x) | 5′-CGCGAGGTTCACACCAAA-3′ 5′-TCCCAAAGTCGTAAGTACAAAATGG-3′ | NM_001135158.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paramonova, I.I.; Vilchinskaya, N.A.; Shenkman, B.S. HDAC4 Is Indispensable for Reduced Slow Myosin Expression at the Early Stage of Hindlimb Unloading in Rat Soleus Muscle. Pharmaceuticals 2021, 14, 1167. https://doi.org/10.3390/ph14111167
Paramonova II, Vilchinskaya NA, Shenkman BS. HDAC4 Is Indispensable for Reduced Slow Myosin Expression at the Early Stage of Hindlimb Unloading in Rat Soleus Muscle. Pharmaceuticals. 2021; 14(11):1167. https://doi.org/10.3390/ph14111167
Chicago/Turabian StyleParamonova, Inna I., Natalia A. Vilchinskaya, and Boris S. Shenkman. 2021. "HDAC4 Is Indispensable for Reduced Slow Myosin Expression at the Early Stage of Hindlimb Unloading in Rat Soleus Muscle" Pharmaceuticals 14, no. 11: 1167. https://doi.org/10.3390/ph14111167
APA StyleParamonova, I. I., Vilchinskaya, N. A., & Shenkman, B. S. (2021). HDAC4 Is Indispensable for Reduced Slow Myosin Expression at the Early Stage of Hindlimb Unloading in Rat Soleus Muscle. Pharmaceuticals, 14(11), 1167. https://doi.org/10.3390/ph14111167







