Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review
Abstract
:1. Introduction
2. Methods
3. Results and Discussion
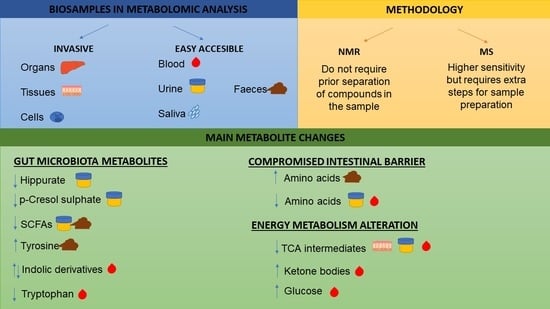
3.1. Biosamples in Metabolomic Analysis
3.2. Methodology to Study the Metabolome of Biological Samples
3.3. Metabolomics Use to Distinguish IBD (CD and UC) Patients from Healthy Controls
3.3.1. Gut Microbiota Metabolites
3.3.2. Metabolic Alterations due to Compromised Intestinal Barrier
3.3.3. Energy Metabolism Alteration
3.4. Metabolomics Use to Distinguish CD from UC, and the Different IBD Subclassifications
3.5. Metabolomic Differences Based on Disease Activity and Predictors of Relapse
3.6. Changes in the Metabolome in Response to Treatment
4. Final Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Burisch, J.; Jess, T.; Martinato, M.; Lakatos, P.L. The burden of inflammatory bowel disease in Europe. J. Crohn’s Colitis 2013, 7, 322–337. [Google Scholar] [CrossRef] [Green Version]
- M’Koma, A.E. Diagnosis of inflammatory bowel disease: Potential role of molecular biometrics. World J. Gastrointest. Surg. 2014, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Chaparro, M.; Zanotti, C.; Burgueño, P.; Vera, I.; Bermejo, F.; Marín-Jiménez, I.; Yela, C.; López, P.; Martín, M.D.; Taxonera, C.; et al. Health care costs of complex perianal fistula in Crohn’s disease. Dig. Dis. Sci. 2013, 58, 3400–3406. [Google Scholar] [CrossRef]
- Fiocchi, C. Integrating omics: The future of IBD? Dig. Dis. 2014, 32, 96–102. [Google Scholar] [CrossRef]
- De Preter, V. Metabolomics in the Clinical Diagnosis of Inflammatory Bowel Disease. Dig. Dis. 2015, 33, 2–10. [Google Scholar] [CrossRef]
- Baldan-Martin, M.; Chaparro, M.; Gisbert, J.P. Tissue Proteomic Approaches to Understand the Pathogenesis of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2021, 27, 1184–1200. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Chaparro, M. Predictors of Primary Response to Biologic Treatment [Anti-TNF, Vedolizumab, and Ustekinumab] in Patients with Inflammatory Bowel Disease: From Basic Science to Clinical Practice. J. Crohn’s Colitis 2020, 14, 694–709. [Google Scholar] [CrossRef]
- Yau, Y.; Leong, R.W.; Zeng, M.; Wasinger, V.C. Proteomics and metabolomics in inflammatory bowel disease. J. Gastroenterol. Hepatol. 2013, 28, 1076–1086. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omic triology. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed. Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Chetwynd, A.J.; Dunn, W.B.; Rodriguez-Blanco, G. Metabolomics: From Fundamentals to Clinical Applications. Adv. Exp. Med. Biol. 2017, 965, 19–44. [Google Scholar]
- Spanogiannopoulos, P.; Bess, E.N.; Carmody, R.N.; Turnbaugh, P.J. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016, 14, 273–287. [Google Scholar] [CrossRef]
- Lenz, E.M.; Bright, J.; Wilson, I.D.; Morgan, S.R.; Nash, A.F.P. A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J. Pharm. Biomed. Anal. 2003, 33, 1103–1115. [Google Scholar] [CrossRef]
- Bezabeh, T.; Somorjai, R.L.; Smith, I.C.P.; Nikulin, A.E.; Dolenko, B.; Bernstein, C.N. The use of 1H magnetic resonance spectroscopy in inflammatory bowel diseases: Distinguishing ulcerative colitis from Crohn’s disease. Am. J. Gastroenterol. 2001, 96, 442–448. [Google Scholar] [CrossRef]
- Balasubramanian, K.; Kumar, S.; Singh, R.R.; Sharma, U.; Ahuja, V.; Makharia, G.K.; Jagannathan, N.R. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: An in vitro proton magnetic resonance spectroscopy study. Magn. Reson. Imaging 2009, 27, 79–86. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Nielsen, O.H.; Hao, F.; Tang, H.; Nicholson, J.K.; Wang, Y.; Olsen, J. Metabonomics in ulcerative colitis: Diagnostics, biomarker identification, and insight into the pathophysiology. J. Proteome Res. 2010, 9, 954–962. [Google Scholar] [CrossRef]
- Sharma, U.; Singh, R.R.; Ahuja, V.; Makharia, G.K.; Jagannathan, N.R. Similarity in the metabolic profile in macroscopically involved and un-involved colonic mucosa in patients with inflammatory bowel disease: An in vitro proton (1H) MR spectroscopy study. Magn. Reson. Imaging 2010, 28, 1022–1029. [Google Scholar] [CrossRef]
- Ooi, M.; Nishiumi, S.; Yoshie, T.; Shiomi, Y.; Kohashi, M.; Fukunaga, K.; Nakamura, S.; Matsumoto, T.; Hatano, N.; Shinohara, M.; et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm. Res. 2011, 60, 831–840. [Google Scholar] [CrossRef]
- Rantalainen, M.; Bjerrum, J.T.; Olsen, J.; Nielsen, O.H.; Wang, Y. Integrative transcriptomic and metabonomic molecular profiling of colonic mucosal biopsies indicates a unique molecular phenotype for ulcerative colitis. J. Proteome Res. 2015, 14, 479–490. [Google Scholar] [CrossRef]
- Keshteli, A.H.; Tso, R.; Dieleman, L.A.; Park, H.; Kroeker, K.I.; Jovel, J.; Gillevet, P.M.; Sikaroodi, M.; Mandal, R.; Fedorak, R.N.; et al. A Distinctive Urinary Metabolomic Fingerprint Is Linked with Endoscopic Postoperative Disease Recurrence in Crohn’s Disease Patients. Inflamm. Bowel Dis. 2018, 24, 861–870. [Google Scholar] [CrossRef]
- Diab, J.; Hansen, T.; Goll, R.; Stenlund, H.; Jensen, E.; Moritz, T.; Florholmen, J.; Forsdahl, G. Mucosal metabolomic profiling and pathway analysis reveal the metabolic signature of ulcerative colitis. Metabolites 2019, 9, 291. [Google Scholar] [CrossRef] [Green Version]
- Santoru, M.L.; Piras, C.; Murgia, F.; Leoni, V.P.; Spada, M.; Murgia, A.; Liggi, S.; Lai, M.A.; Usai, P.; Caboni, P.; et al. Metabolic Alteration in Plasma and Biopsies from Patients with IBD. Inflamm. Bowel Dis. 2021, 27, 1335–1345. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Steenholdt, C.; Ainsworth, M.; Nielsen, O.H.; Reed, M.A.C.; Atkins, K.; Günther, U.L.; Hao, F.; Wang, Y. Metabonomics uncovers a reversible proatherogenic lipid profile during infliximab therapy of inflammatory bowel disease. BMC Med. 2017, 15, 184. [Google Scholar] [CrossRef] [Green Version]
- Murgia, A.; Hinz, C.; Liggi, S.; Denes, J.; Hall, Z.; West, J.; Santoru, M.L.; Piras, C.; Manis, C.; Usai, P.; et al. Italian cohort of patients affected by inflammatory bowel disease is characterised by variation in glycerophospholipid, free fatty acids and amino acid levels. Metabolomics 2018, 14, 140. [Google Scholar] [CrossRef]
- Scoville, E.A.; Allaman, M.M.; Brown, C.T.; Motley, K.; Horst, S.N.; Williams, C.S.; Koyama, T.; Zhao, Z.; Adams, D.W.; Beaulieu, D.B.; et al. Alterations in lipid, amino acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics 2018, 14, 17. [Google Scholar] [CrossRef]
- Probert, F.; Walsh, A.; Jagielowicz, M.; Yeo, T.; Claridge, T.D.W.; Simmons, A.; Travis, S.; Anthony, D.C. Plasma nuclear magnetic resonance metabolomics discriminates between high and low endoscopic activity and predicts progression in a prospective cohort of patients with ulcerative colitis. J. Crohn’s Colitis 2018, 12, 1326–1337. [Google Scholar] [CrossRef]
- Notararigo, S.; Martín-Pastor, M.; Viñuela-Roldán, J.E.; Quiroga, A.; Dominguez-Munoz, J.E.; Barreiro-de Acosta, M. Targeted 1H NMR metabolomics and immunological phenotyping of human fresh blood and serum samples discriminate between healthy individuals and inflammatory bowel disease patients treated with anti-TNF. J. Mol. Med. 2021, 99, 1251–1264. [Google Scholar] [CrossRef]
- Sofia, M.A.; Ciorba, M.A.; Meckel, K.; Lim, C.K.; Guillemin, G.J.; Weber, C.R.; Bissonnette, M.; Pekow, J.R. Tryptophan metabolism through the kynurenine pathway is associated with endoscopic inflammation in ulcerative colitis. Inflamm. Bowel Dis. 2018, 24, 1471–1480. [Google Scholar] [CrossRef]
- Lai, Y.; Xue, J.; Liu, C.W.; Gao, B.; Chi, L.; Tu, P.; Lu, K.; Ru, H. Serum metabolomics identifies altered bioenergetics, signaling cascades in parallel with exposome markers in Crohn’s disease. Molecules 2019, 24, 449. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Du, B.; Shi, Y.; Lu, Y.; Zhou, Y.; Liu, B. Combined signature of the fecal microbiome and plasma metabolome in patients with ulcerative colitis. Med. Sci. Monit. 2019, 25, 3303–3315. [Google Scholar] [CrossRef]
- Roda, G.; Porru, E.; Katsanos, K.; Skamnelos, A.; Kyriakidi, K.; Fiorino, G.; Christodoulou, D.; Danese, S.; Roda, A. Serum Bile Acids Profiling in Inflammatory Bowel Disease Patients Treated with Anti-TNFs. Cells 2019, 8, 817. [Google Scholar] [CrossRef] [Green Version]
- Krzystek-Korpacka, M.; Fleszar, M.G.; Bednarz-misa, I.; Lewandowski, Ł.; Szczuka, I.; Kempiński, R.; Neubauer, K. Transcriptional and metabolomic analysis of l-arginine/nitric oxide pathway in inflammatory bowel disease and its association with local inflammatory and angiogenic response: Preliminary findings. Int. J. Mol. Sci. 2020, 21, 1641. [Google Scholar] [CrossRef] [Green Version]
- Williams, H.R.T.; Willsmore, J.D.; Cox, I.J.; Walker, D.G.; Cobbold, J.F.L.; Taylor-Robinson, S.D.; Orchard, T.R. Serum metabolic profiling in inflammatory bowel disease. Dig. Dis. Sci. 2012, 57, 2157–2165. [Google Scholar] [CrossRef]
- Di Giovanni, N.; Meuwis, M.A.; Louis, E.; Focant, J.F. Untargeted Serum Metabolic Profiling by Comprehensive Two-Dimensional Gas Chromatography-High-Resolution Time-of-Flight Mass Spectrometry. J. Proteome Res. 2020, 19, 1013–1028. [Google Scholar] [CrossRef]
- Borren, N.Z.; Plichta, D.; Joshi, A.D.; Bonilla, G.; Peng, V.; Colizzo, F.P.; Luther, J.; Khalili, H.; Garber, J.J.; van der Woude, C.J.; et al. Alterations in Fecal Microbiomes and Serum Metabolomes of Fatigued Patients with Quiescent Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2021, 19, 519–527.e5. [Google Scholar] [CrossRef]
- Ding, N.S.; McDonald, J.A.K.; Perdones-Montero, A.; Rees, D.N.; Adegbola, S.O.; Misra, R.; Hendy, P.; Penez, L.; Marchesi, J.R.; Holmes, E.; et al. Metabonomics and the Gut Microbiome Associated With Primary Response to Anti-TNF Therapy in Crohn’s Disease. J. Crohn’s Colitis 2020, 14, 1090–1102. [Google Scholar] [CrossRef]
- Borren, N.Z.; Plichta, D.; Joshi, A.D.; Bonilla, G.; Sadreyev, R.; Vlamakis, H.; Xavier, R.J.; Ananthakrishnan, A.N. Multi-“-Omics” Profiling in Patients with Quiescent Inflammatory Bowel Disease Identifies Biomarkers Predicting Relapse. Inflamm. Bowel Dis. 2020, 26, 1524–1532. [Google Scholar] [CrossRef]
- Tefas, C.; Ciobanu, L.; Tanțău, M.; Moraru, C.; Socaciu, C. The potential of metabolic and lipid profiling in inflammatory bowel diseases: A pilot study. Bosn. J. Basic Med. Sci. 2020, 20, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative metabolomic profiling of serum, plasma, and urine by 1H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, L.; Xu, Y.; Lin, Y.; Jin, Y.; Zheng, C. 1H NMR-based spectroscopy detects metabolic alterations in serum of patients with early-stage ulcerative colitis. Biochem. Biophys. Res. Commun. 2013, 433, 547–551. [Google Scholar] [CrossRef]
- Dawiskiba, T.; Deja, S.; Mulak, A.; Zabek, A.; Jawień, E.; Pawełka, D.; Banasik, M.; Mastalerz-Migas, A.; Balcerzak, W.; Kaliszewski, K.; et al. Serum and urine metabolomic fngerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 163–174. [Google Scholar] [CrossRef]
- Hisamatsu, T.; Ono, N.; Imaizumi, A.; Mori, M.; Suzuki, H.; Uo, M.; Hashimoto, M.; Naganuma, M.; Matsuoka, K.; Mizuno, S.; et al. Decreased plasma histidine level predicts risk of relapse in patients with ulcerative colitis in remission. PLoS ONE 2015, 10, e0140716. [Google Scholar]
- Wilson, A.; Teft, W.A.; Morse, B.L.; Choi, Y.H.; Woolsey, S.; DeGorter, M.K.; Hegele, R.A.; Tirona, R.G.; Kim, R.B. Trimethylamine-N-oxide: A Novel Biomarker for the Identification of Inflammatory Bowel Disease. Dig. Dis. Sci. 2015, 60, 3620–3630. [Google Scholar] [CrossRef]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516.e2. [Google Scholar] [CrossRef] [Green Version]
- Keshteli, A.H.; Den Brand, F.F.V.; Madsen, K.L.; Mandal, R.; Valcheva, R.; Kroeker, K.I.; Han, B.; Bell, R.C.; Cole, J.; Hoevers, T.; et al. Dietary and metabolomic determinants of relapse in ulcerative colitis patients: A pilot prospective cohort study. World J. Gastroenterol. 2017, 23, 3890–3899. [Google Scholar] [CrossRef]
- Williams, H.R.T.; Cox, I.J.; Walker, D.G.; North, B.V.; Patel, V.M.; Marshall, S.E.; Jewell, D.P.; Ghosh, S.; Thomas, H.J.W.; Teare, J.P.; et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am. J. Gastroenterol. 2009, 104, 1435–1444. [Google Scholar] [CrossRef]
- Stephens, N.S.; Siffledeen, J.; Su, X.; Murdoch, T.B.; Fedorak, R.N.; Slupsky, C.M. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J. Crohn’s Colitis 2013, 7, e42–e48. [Google Scholar] [CrossRef] [Green Version]
- Alonso, A.; Julià, A.; Vinaixa, M.; Domènech, E.; Fernández-Nebro, A.; Cañete, J.D.; Ferrándiz, C.; Tornero, J.; Gisbert, J.P.; Nos, P.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 133. [Google Scholar] [CrossRef]
- Piestansky, J.; Olesova, D.; Galba, J.; Marakova, K.; Parrak, V.; Secnik, P.; Secnik, P.; Kovacech, B.; Kovac, A.; Zelinkova, Z.; et al. Profiling of amino acids in urine samples of patients suffering from inflammatory bowel disease by capillary electrophoresis-mass spectrometry. Molecules 2019, 24, 3345. [Google Scholar] [CrossRef] [Green Version]
- Keshteli, A.H.; Madsen, K.L.; Mandal, R.; Boeckxstaens, G.E.; Bercik, P.; De Palma, G.; Reed, D.E.; Wishart, D.; Vanner, S.; Dieleman, L.A. Comparison of the metabolomic profiles of irritable bowel syndrome patients with ulcerative colitis patients and healthy controls: New insights into pathophysiology and potential biomarkers. Aliment. Pharmacol. Ther. 2019, 49, 723–732. [Google Scholar] [CrossRef]
- Fang, X.; Vázquez-Baeza, Y.; Elijah, E.; Vargas, F.; Ackermann, G.; Humphrey, G.; Lau, R.; Weldon, K.C.; Sanders, J.G.; Panitchpakdi, M.; et al. Gastrointestinal Surgery for Inflammatory Bowel Disease Persistently Lowers Microbiome and Metabolome Diversity. Inflamm. Bowel Dis. 2021, 27, 603–616. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and Noninvasive Metabonomic Characterization of Inflammatory Bowel Disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef]
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE 2009, 4, e6386. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Wang, Y.; Hao, F.; Coskun, M.; Ludwig, C.; Günther, U.; Nielsen, O.H. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics 2014, 11, 122–133. [Google Scholar] [CrossRef] [Green Version]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-madi, A.; Avila-pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; Mciver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- de Freitas Lins Neto, M.Á.; Verdi, G.M.X.; de Oliveira Veras, A.; de Oliveira Veras, M.; Caetano, L.C.; Ursulino, J.S. Use of metabolomics to the diagnosis of inflammatory bowel disease. Arq. Gastroenterol. 2020, 57, 311–315. [Google Scholar] [CrossRef]
- David, A.; Rostkowski, P. Analytical Techniques in Metabolomics; Elsevier Inc.: Amsterdam, The Netherlands, 2020; ISBN 9780128181966. [Google Scholar]
- Crook, A.A.; Powers, R. Quantitative NMR-Based Biomedical Metabolomics: Current Status and Applications. Molecules 2020, 25, 5128. [Google Scholar] [CrossRef]
- Zia, K.; Siddiqui, T.; Ali, S.; Farooq, I.; Zafar, M.S.; Khurshid, Z. Nuclear Magnetic Resonance Spectroscopy for Medical and Dental Applications: A Comprehensive Review. Eur. J. Dent. 2019, 13, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-H.; Lee, B.-Y.; Tuite, P.; Coles, L.; Sathe, A.G.; Chen, C.; Cloyd, J.; Low, W.C.; Steer, C.J.; Chen, W. Quantitative Assessment of Occipital Metabolic and Energetic Changes in Parkinson’s Patients, Using In Vivo 31P MRS-Based Metabolic Imaging at 7T. Metabolites 2021, 11, 145. [Google Scholar] [CrossRef]
- Filzmoser, P.; Walczak, B. What can go wrong at the data normalization step for identification of biomarkers? J. Chromatogr. A 2014, 1362, 194–205. [Google Scholar] [CrossRef]
- Gallagher, K.; Catesson, A.; Griffin, J.L.; Holmes, E.; Williams, H.R.T. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J. Crohn’s Colitis 2021, 15, 813–826. [Google Scholar] [CrossRef]
- Aldars-García, L.; Marin, A.C.; Chaparro, M.; Gisbert, J.P. The interplay between immune system and microbiota in inflammatory bowel disease: A narrative review. Int. J. Mol. Sci. 2021, 22, 3076. [Google Scholar] [CrossRef]
- Yap, I.K.S.; Li, J.V.; Saric, J.; Martin, F.P.; Davies, H.; Wang, Y.; Wilson, I.D.; Nicholson, J.K.; Utzinger, J.; Marchesi, J.R.; et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-lnduced gut microbiota modification in the mouse. J. Proteome Res. 2008, 7, 3718–3728. [Google Scholar] [CrossRef] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Walker, A.; Schmitt-Kopplin, P. The role of fecal sulfur metabolome in inflammatory bowel diseases. Int. J. Med. Microbiol. 2021, 311, 151513. [Google Scholar] [CrossRef]
- Herrema, H.; Niess, J.H. Intestinal microbial metabolites in human metabolism and type 2 diabetes. Diabetologia 2020, 63, 2533–2547. [Google Scholar] [CrossRef]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic review: The gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef]
- Lanis, J.M.; Alexeev, E.E.; Curtis, V.F.; Kitzenberg, D.A.; Kao, D.J.; Battista, K.D.; Gerich, M.E.; Glover, L.E.; Kominsky, D.J.; Colgan, S.P. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017, 10, 1133–1144. [Google Scholar] [CrossRef]
- Grifka-Walk, H.M.; Jenkins, B.R.; Kominsky, D.J. Amino Acid Trp: The Far Out Impacts of Host and Commensal Tryptophan Metabolism. Front. Immunol. 2021, 12, 653208. [Google Scholar] [CrossRef]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and Signaling Pathways of Amino Acids in Intestinal Inflammation. Biomed Res. Int. 2018, 9171905. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [Green Version]
| Category | No of Studies | References | ||
|---|---|---|---|---|
| Biosamples in Metabolomic Analysis | Tissue | 9 | [14,15,16,17,18,19,20,21,22] | |
| Blood | 25 | [18,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45] | ||
| Urine | 10 | [16,36,39,41,45,46,47,48,49,50] | ||
| Faeces | 11 | [30,36,51,52,53,54,55,56,57,58,59] | ||
| Methodology | NMR | 21 | [14,15,16,17,19,20,23,26,27,33,39,40,41,46,47,48,52,54,55,59] | |
| MS | 26 | [18,20,21,22,24,25,28,29,30,31,32,34,37,38,42,43,44,45,49,50,51,56,57,58] | ||
| Main Metabolite Changes | Gut Microbiota Metabolites | Decrease in urinary hippurate | 5 | [39,41,46,47,48] |
| Decrease in urinary p-cresol sulphate | 2 | [41,46] | ||
| Decrease in urinary and faecal SCFAs | 10 | [30,39,41,47,48,52,54,55,57,58] | ||
| Increase in faecal tyrosine | 4 | [30,53,55,59] | ||
| Decrease in serological/plasmatic tryptophan | 7 | [18,24,27,29,35,41,44] | ||
| Compromised Intestinal Barrier | Increase in faecal amino acids | 8 | [30,52,53,54,55,56,59] | |
| Decrease in urinary and serological/plasmatic amino acids | 19 | [16,18,23,24,25,27,29,32,33,34,35,36,39,40,42,45,47,48,49] | ||
| Energy Metabolism Alteration | Decrease in serological/plasmatic, urinary and tissular TCA intermediates | 11 | [15,18,23,25,34,36,39,40,41,47,48] | |
| Increase in serological/plasmatic ketone bodies | 7 | [18,33,34,39,40,41,45] | ||
| Increase in serological/plasmatic glucose | 5 | [26,34,39,40,41] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldars-García, L.; Gisbert, J.P.; Chaparro, M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals 2021, 14, 1190. https://doi.org/10.3390/ph14111190
Aldars-García L, Gisbert JP, Chaparro M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals. 2021; 14(11):1190. https://doi.org/10.3390/ph14111190
Chicago/Turabian StyleAldars-García, Laila, Javier P. Gisbert, and María Chaparro. 2021. "Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review" Pharmaceuticals 14, no. 11: 1190. https://doi.org/10.3390/ph14111190
APA StyleAldars-García, L., Gisbert, J. P., & Chaparro, M. (2021). Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals, 14(11), 1190. https://doi.org/10.3390/ph14111190