Biostimulation of Bacteria in Liquid Culture for Identification of New Antimicrobial Compounds
Abstract
:1. Introduction
2. Results
2.1. Culture-Dependent Plate Assay and Identification of Bacterial Isolates
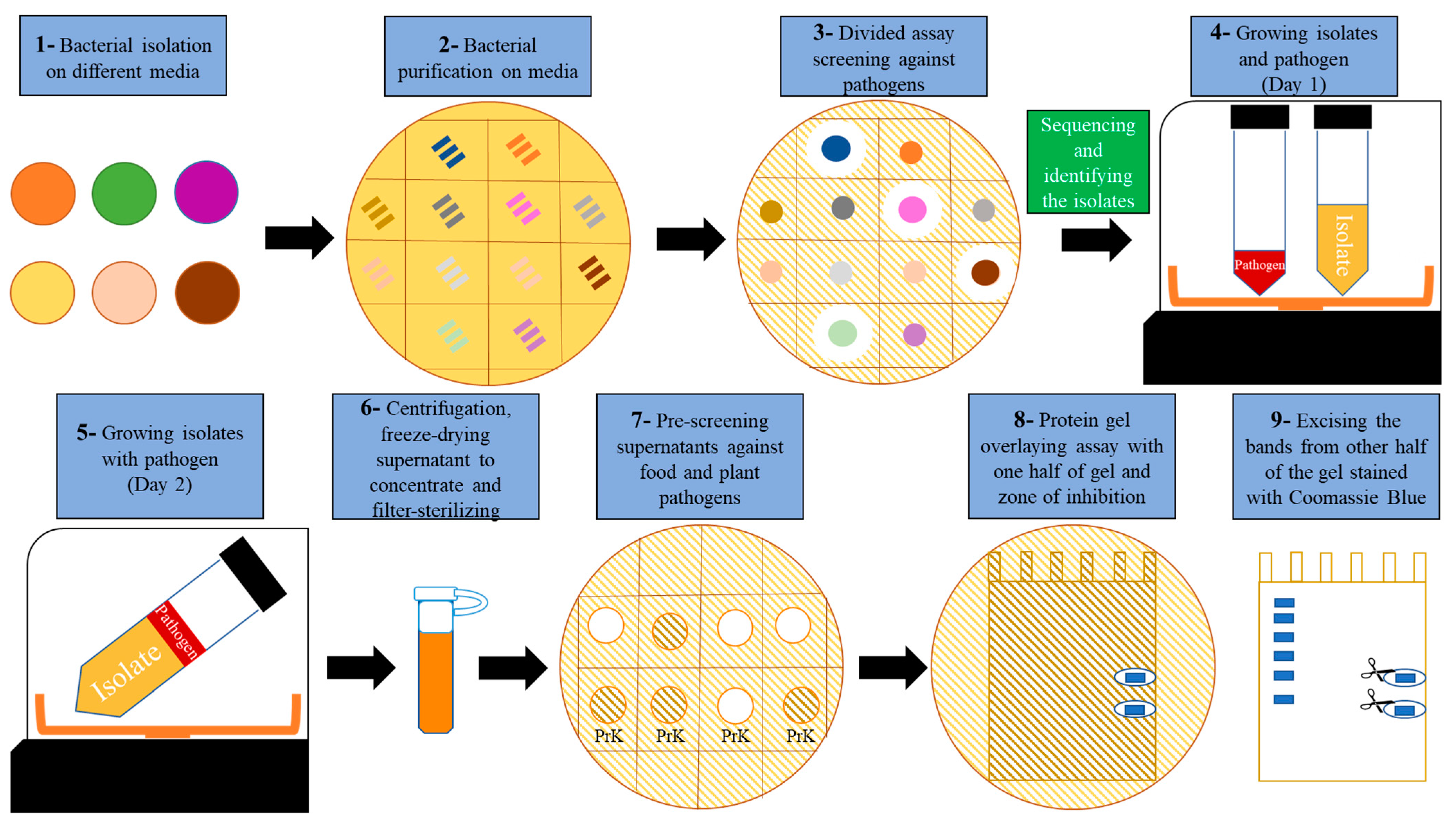
2.2. Development of an Antimicrobial Discovery Pipeline
2.3. Stimulation of AMC Production by Exposure to Living Plant Pathogens
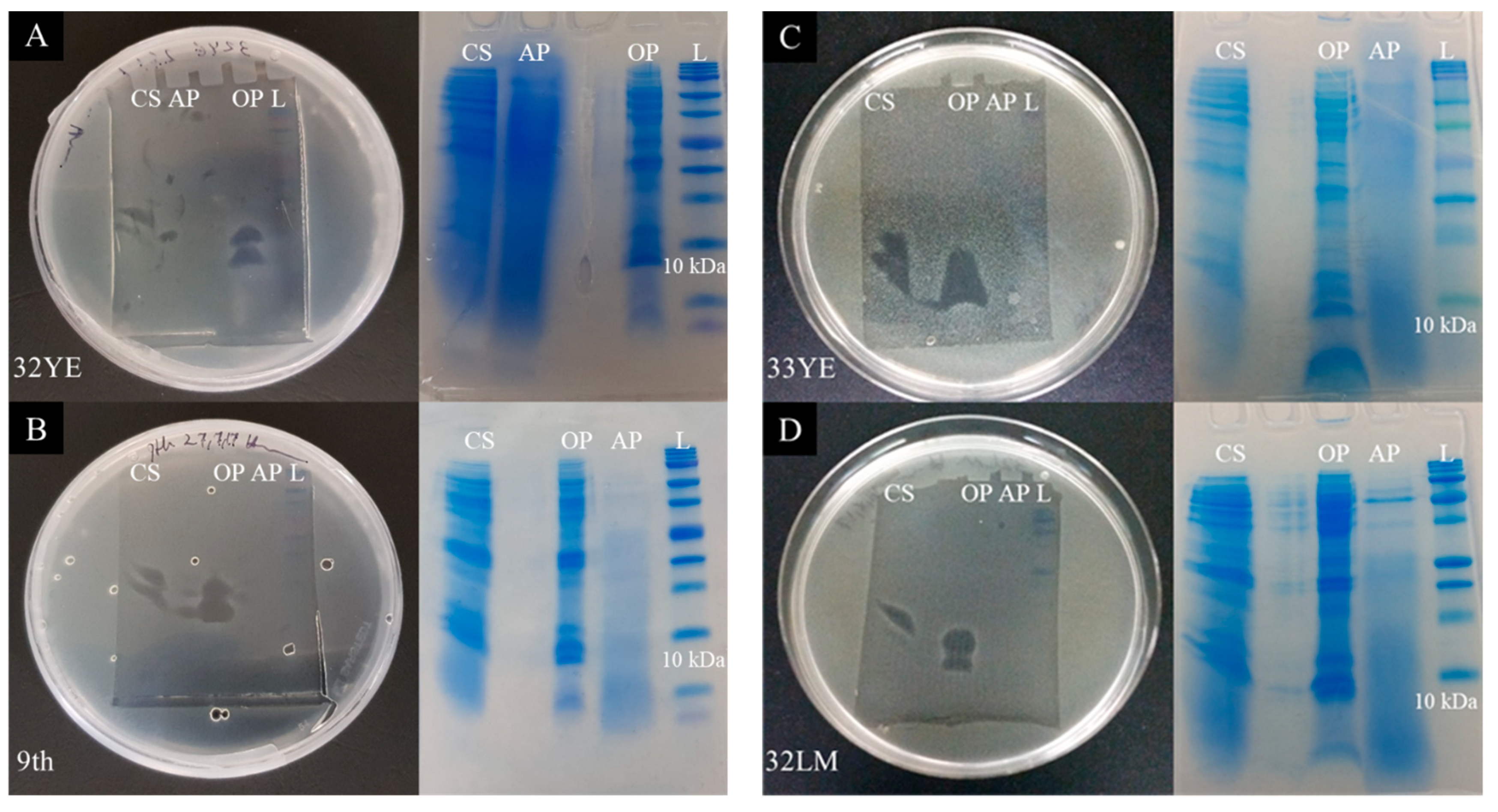
2.4. Peptide Gel Microbial Inhibition Assay and Partial Identification of Antimicrobial Peptides
2.5. Larger Proteins
2.6. Identification of Small Antimicrobial Peptides through HPLC Fractionation
2.7. Identification of Antimicrobial Metabolites
3. Discussion
4. Materials and Methods
4.1. Isolation of Bacteria from Soil and Food Spoilage Material Using Different Media
4.2. Plate Pathogen Inhibition Assay
4.3. Identifying Potential Biocontrol Bacteria
4.4. AMC Induction Assay
4.5. Antibacterial and Peptidase Assays
4.6. Peptide Gel Microbial Inhibition Assay
4.7. Partial Characterization of Antimicrobial Peptides by Tandem Mass Spectrometry
4.8. Amino Acid Data Analysis
4.9. HPLC (RP-HPLC)
4.10. GC/MS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Castro, A.; Silva, J.; Teixeira, P. Staphylococcus aureus, a food pathogen: Virulence factors and antibiotic resistance. In Foodborne Diseases; Academic Press: London, UK, 2018; pp. 213–238. [Google Scholar]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Hug, L.A.; Baker, B.; Anantharaman, K.; Brown, C.T.; Probst, A.; Castelle, C.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Pliego, C.; Ramos, C.; de Vicente, A.; Cazorla, F.M. Screening for candidate bacterial biocontrol agents against soilborne fungal plant pathogens. Plant Soil 2010, 340, 505–520. [Google Scholar] [CrossRef] [Green Version]
- Schmeisser, C.; Steele, H.; Streit, W.R. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 2007, 75, 955–962. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Genet. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial production of bacteriocins: Latest research development and applications. Biotechnol. Adv. 2018, 36, 2187–2200. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Genet. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Garbeva, P.; Silby, M.W.; Raaijmakers, J.; Levy, S.B.; de Boer, W. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J. 2011, 5, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Tyc, O.; Berg, M.V.D.; Gerards, S.; Van Veen, J.A.; Raaijmakers, J.; de Boer, W.; Garbeva, P. Impact of interspecific interactions on antimicrobial activity among soil bacteria. Front. Microbiol. 2014, 5, 567. [Google Scholar] [CrossRef]
- Kim, J.; Yoon, S.; Park, Y.; Kim, S.; Ryu, C. Crossing the kingdom border: Human diseases caused by plant pathogens. Environ. Microbiol. 2020, 22, 2485–2495. [Google Scholar] [CrossRef]
- Garbeva, P.; Tyc, O.; Remus-Emsermann, M.; van der Wal, A.; Vos, M.; Silby, M.; de Boer, W. No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1. PLoS ONE 2011, 6, e27266. [Google Scholar] [CrossRef]
- Makovitzki, A.; Shai, Y. pH-dependent antifungal lipopeptides and their plausible mode of action. Biochemistry 2005, 44, 9775–9784. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, X.; Pan, J.; Su, X.; Jin, X.; Guan, X. Purification and characterization of a novel cold shock protein-like bacteriocin synthesized by Bacillus thuringiensis. Sci. Rep. 2016, 6, 35560. [Google Scholar] [CrossRef] [Green Version]
- Nordberg, J.; Arnér, E.S. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001, 31, 1287–1312. [Google Scholar] [CrossRef]
- Mostertz, J.; Hochgräfe, F.; Jürgen, B.; Schweder, T.; Hecker, M. The role of thioredoxin TrxA in Bacillus subtilis: A proteomics and transcriptomics approach. Proteomics 2008, 8, 2676–2690. [Google Scholar] [CrossRef]
- Park, S.-C.; Jung, Y.J.; Kim, I.R.; Lee, J.R.; Jang, M.-K. Functional characterization of thioredoxin h type 5 with antimicrobial activity from Arabidopsis thaliana. Biotechnol. Bioprocess Eng. 2017, 22, 129–135. [Google Scholar] [CrossRef]
- Saibil, H.R. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goltermann, L.; Sarusie, M.V.; Ebentin, T. Chaperonin GroEL/GroES over-expression promotes aminoglycoside resistance and reduces drug susceptibilities in Escherichia coli following exposure to sublethal aminoglycoside doses. Front. Microbiol. 2016, 6, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, A.; Derkaoui, M.; Terrade, A.; Denizon, M.; Deghmane, A.-E.; Deutscher, J.; Delany, I.; Taha, M.-K. The phosphocarrier protein HPr contributes to meningococcal survival during infection. PLoS ONE 2016, 11, e0162434. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.; Portnoy, D.A. SpoVG is a conserved RNA-binding protein that regulates Listeria monocytogenes lysozyme resistance, virulence, and swarming motility. mBio 2016, 7, e00240-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Devine, R.; Hutchings, M.I.; Wilkinson, B. A role for antibiotic biosynthesis monooxygenase domain proteins in fidelity control during aromatic polyketide biosynthesis. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, V.; Iqbal, A.M.Z.; Haseeb, M.; Khan, M.S. Antimicrobial potential of bacteriocin producing Lysinibacillus jx416856 against foodborne bacterial and fungal pathogens, isolated from fruits and vegetable waste. Anaerobe 2014, 27, 87–95. [Google Scholar] [CrossRef]
- Pidutti, P.; Federici, F.; Brandi, J.; Manna, L.; Rizzi, E.; Marini, U.; Cecconi, D. Purification and characterization of ribosomal proteins L27 and L30 having antimicrobial activity produced by the Lactobacillus salivarius SGL 03. J. Appl. Microbiol. 2018, 124, 398–407. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Fraise, A.; Wilkinson, M.; Bradley, C.; Oppenheim, B.; Moiemen, N. The antibacterial activity and stability of acetic acid. J. Hosp. Infect. 2013, 84, 329–331. [Google Scholar] [CrossRef]
- Basch, H.; Gadebusch, H. In vitro antimicrobial activity of dimethylsulfoxide. Appl. Microbiol. 1968, 16, 1953. [Google Scholar] [CrossRef]
- Oh, D.-H.; Marshall, D.L. Antimicrobial activity of ethanol, glycerol monolaurate or lactic acid against Listeria monocytogenes. Int. J. Food Microbiol. 1993, 20, 239–246. [Google Scholar] [CrossRef]
- Yi, H.-S.; Ahn, Y.-R.; Song, G.C.; Ghim, S.-Y.; Lee, S.; Lee, G.; Ryu, C.-M. Impact of a bacterial volatile 2,3-butanediol on Bacillus subtilis rhizosphere robustness. Front. Microbiol. 2016, 7, 993. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, M.; Abraham, W.-R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. [Google Scholar] [CrossRef] [PubMed]
- Kadaikunnan, S.; Rejiniemon, T.S.; Khaled, J.M.; Alharbi, N.S.; Mothana, R. In vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kiran, G.S.; Priyadharsini, S.; Sajayan, A.; Ravindran, A.; Selvin, J. An antibiotic agent pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro isolated from a marine bacteria Bacillus tequilensis MSI45 effectively controls multi-drug resistant Staphylococcus aureus. RSC Adv. 2018, 8, 17837–17846. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.-L.; Han, T.; Wu, J.-Z.; Zhang, Q.-Y.; Zhang, H.; Huang, B.-K.; Rahman, K.; Qin, L.-P. Comparative research of chemical constituents, antifungal and antitumor properties of ether extracts of Panax ginseng and its endophytic fungus. Phytomedicine 2009, 16, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Tonial, F.; Maia, B.H.L.N.S.; Gomes-Figueiredo, J.A.; Sobottka, A.M.; Bertol, C.D.; Nepel, A.; Savi, D.C.; Vicente, V.; Gomes, R.R.; Glienke, C. Influence of culturing conditions on bioprospecting and the antimicrobial potential of endophytic fungi from Schinus terebinthifolius. Curr. Microbiol. 2015, 72, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Carrión, O.; Curson, A.R.J.; Kumaresan, D.; Fu, Y.; Lang, A.; Mercade, E.; Todd, J. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bamforth, C.W. Dimethyl sulfide–significance, origins, and control. J. Am. Soc. Brew. Chem. 2014, 72, 165–168. [Google Scholar] [CrossRef] [Green Version]
- Nandi, D.L.; Horowitz, P.M.; Westley, J. Rhodanese as a thioredoxin oxidase. Int. J. Biochem. Cell Biol. 2000, 32, 465–473. [Google Scholar] [CrossRef]
- Nagahara, N. Catalytic site cysteines of thiol enzyme: Sulfurtransferases. J. Amino Acids 2011, 2011, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Martins, M.B.; Carvalho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923–9932. [Google Scholar] [CrossRef]
- Giessen, T.W.; Marahiel, M.A. Rational and combinatorial tailoring of bioactive cyclic dipeptides. Front. Microbiol. 2015, 6, 785. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of lake Chaka, an athalassohaline lake in Northwestern China. Appl. Environ. Microbiol. 2006, 72, 3832–3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Isolates Name | Isolates’ LAB ID | C. michiganensis | L. monocytogenes | P. syringae | E. coli |
|---|---|---|---|---|---|
| B. pumilus | 18M | AMM | AMM | ||
| 20M | AMP | ||||
| 44YE | AMM | AMP | |||
| 8LM | AMP | ||||
| 32LM | AMP | ||||
| 33LM | AMP | ||||
| 35LM | AMP | ||||
| B. subtilis | 1M1 | AMP | AMP | ||
| 25LGS | AMP | AMP | |||
| B. amyloliquefaciens | 33YE | AMM | AMP | ||
| B. licheniformis | 28M | AMP | |||
| B. mojavensis | 30LM | AMP | |||
| B. circulans | 9th | AMP | |||
| B. methylotrophicus | 46YE | AMP | |||
| 45YE | AMP | ||||
| 42LGS | AMM | AMM | |||
| B. safensis | 35YE | AMM | AMP | ||
| 10th | AMP | ||||
| B. laterosporus | 4YE | AMM | AMP | ||
| P. peoriae/polymyxa | 14th | AMM | AMM | ||
| L. fusiformis | 34LM | AMM | |||
| L. mangiferihumi | 37LM | AMP | AMM | ||
| K. pneumoniae | 44LGF | AMP | |||
| S. aquimarina | 32YE | AMP | AMP | ||
| 19YE | AMP | AMP | |||
| S. saromensis | 39YE | AMP | AMP | ||
| After adjustment to pH 7 | |||||
| S. saprophyticus/xylosus | 36M | AMP | AMM | ||
| 41M | AMP | AMM | |||
| S. sciuri | 42M | AMP | AMM | ||
| P. vulgaris | 30MC | AMP/AMM | |||
| E. ludwigii | 46M | AMM | |||
| E. cloacae | 42MC | AMP/AMM | |||
| C. jiangduensis | 28MC | AMP/AMM | |||
| C. flavescens | 43M | AMM | AMM | ||
| O. grignonense/pseudogrignonense | 34MC | AMP | |||
| 37MC | AMP | ||||
| Genera | Cold-Shock Protein | Thioredoxin (TRX) | DNA-Binding Protein | RNA-Binding Protein | Phosphocarrier Protein HPr | 10 kDa Co-Chaperone (Co-Chaperonin GroES) | Septation Protein SpoVG | 30S Ribosomal Proteins | 50S Ribosomal Proteins | Hypothetical Proteins | Antibiotic Biosynthesis monooxygenase |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus spp. (9th, 10th, 32LM, 33LM, 35LM, 33YE, 35YE, 44 YE, 45YE, 46YE) | ✓ | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | - |
| Lysinibacillus spp. (37LM) | - | ✓ | - | ✓ | - | - | ✓ | - | - | ✓ | ✓ |
| Sporosarcina spp. (32YE and 39YE) | ✓ | ✓ | ✓ | - | - | - | ✓ | - | ✓ | - | - |
| Brevibacillus spp. (4YE) | ✓ | - | - | - | - | - | - | - | ✓ | - | - |
| Name of Isolates | Potential AMMs |
|---|---|
| Bacillus spp. | 2,3-Butanediol |
| 30LM, 32LM, 35LM | Acetic acid |
| 35YE, 45YE, 46YE, | DMSO |
| 18M, 20M, 10th | Glycerol |
| 3-Isobutyl hexahydropyrrolo[1,2-a]pyrazine-1,4-dione *, ** | |
| Dihydro-1H-pyrrolizine-3,5(2H,6H)-dione | |
| (2S)-1-(2-Aminoacetyl)pyrrolidine-2-carboxylic acid | |
| 2,3,6,7,8,8a-Hexahydropyrrolo[1,2-a]pyrazine-1,4-dione | |
| 2-Acetyl-3-methyl-octahydropyrrolo[1,2-a]piperazine-1,4-dione | |
| 1,4,5-Triethyltetrazaborole | |
| 3-Butyl-6-methylpiperazine-2,5-dione | |
| 5-Methylimidazolidine-2,4-dione | |
| Lysinibacillus spp. | 2,3-Butanediol |
| 37LM, 34LM | DMSO |
| Glycerol | |
| 3-Isobutyl hexahydropyrrolo[1,2-a]pyrazine-1,4-dione | |
| 2-Acetyl-3-methyl-octahydropyrrolo[1,2-a]piperazine-1,4-dione | |
| 2,3,6,7,8,8a-Hexahydropyrrolo[1,2-a]pyrazine-1,4-dione | |
| Paenibacillaceae spp. | Acetic acid |
| 4YE, 14th | DMSO |
| 2,3,6,7,8,8a-Hexahydropyrrolo[1,2-a]pyrazine-1,4-dione | |
| 3-Isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione | |
| 3-Butyl-6-methylpiperazine-2,5-dione | |
| Klebsiella spp. | DMSO |
| 44LGF | Dimethyl palmitamine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirzaee, H.; Ariens, E.; Blaskovich, M.A.T.; Clark, R.J.; Schenk, P.M. Biostimulation of Bacteria in Liquid Culture for Identification of New Antimicrobial Compounds. Pharmaceuticals 2021, 14, 1232. https://doi.org/10.3390/ph14121232
Mirzaee H, Ariens E, Blaskovich MAT, Clark RJ, Schenk PM. Biostimulation of Bacteria in Liquid Culture for Identification of New Antimicrobial Compounds. Pharmaceuticals. 2021; 14(12):1232. https://doi.org/10.3390/ph14121232
Chicago/Turabian StyleMirzaee, Hooman, Emily Ariens, Mark A. T. Blaskovich, Richard J. Clark, and Peer M. Schenk. 2021. "Biostimulation of Bacteria in Liquid Culture for Identification of New Antimicrobial Compounds" Pharmaceuticals 14, no. 12: 1232. https://doi.org/10.3390/ph14121232
APA StyleMirzaee, H., Ariens, E., Blaskovich, M. A. T., Clark, R. J., & Schenk, P. M. (2021). Biostimulation of Bacteria in Liquid Culture for Identification of New Antimicrobial Compounds. Pharmaceuticals, 14(12), 1232. https://doi.org/10.3390/ph14121232








