Mechanism of Resistance Development in E. coli against TCAT, a Trimethoprim-Based Photoswitchable Antibiotic
Abstract
:1. Introduction
2. Results
2.1. DHFR Inhibition Assay
2.2. Isolation of Resistant Variants
2.3. Genomic Mutations Cannot Fully Explain the Acquired Resistance
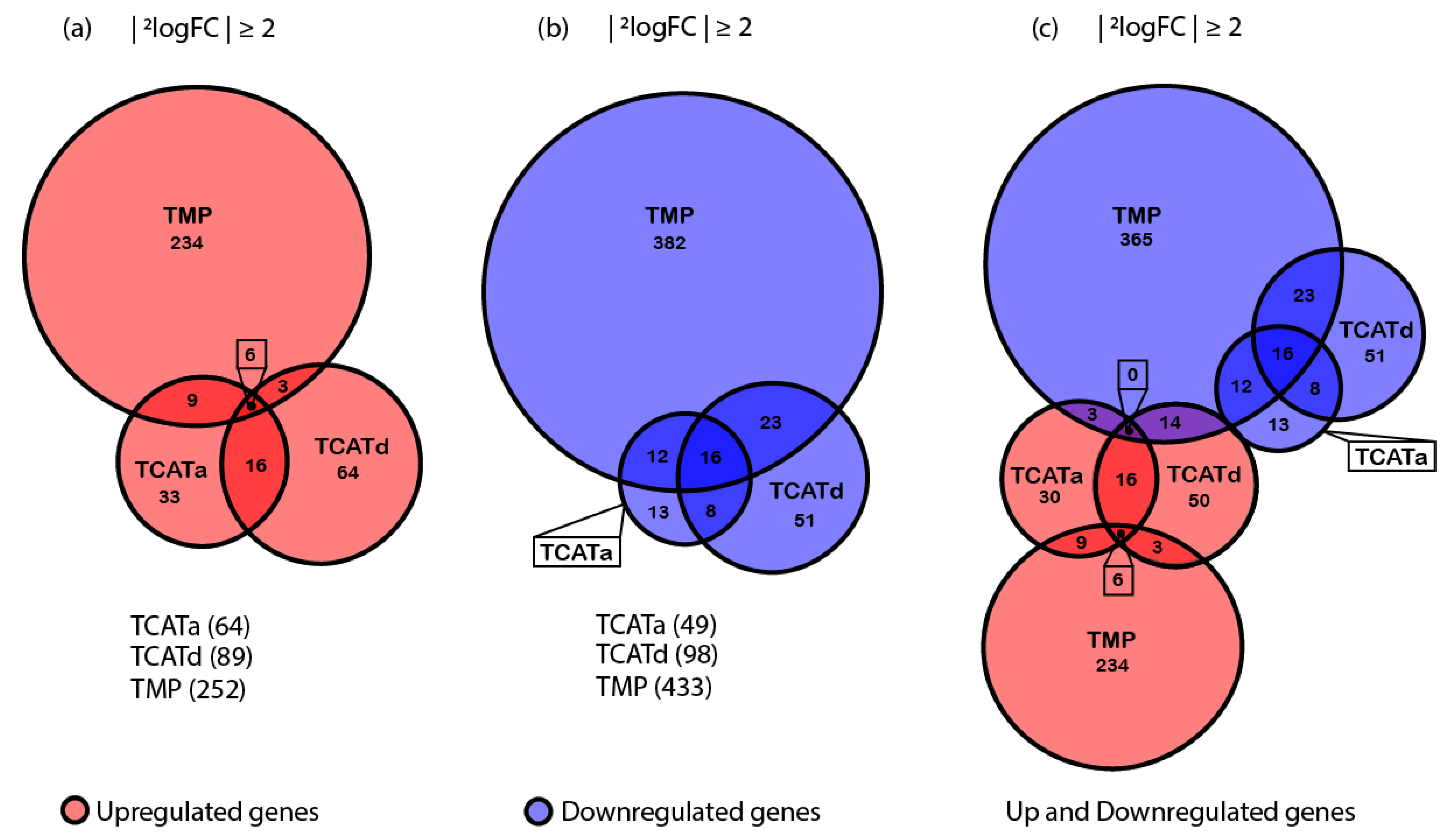
2.4. Transcriptomics Reveals Different Approaches towards Resistance Development between Isolates
2.5. Shared Gene Expression between Isolates Corresponds with a Stress Response and Reduced Cell Growth
2.6. TCATa Induces Expression of Colonic Acid, Exopolysaccharides and TolC
2.7. TCATd Activates Carnitine Synthesis, Maltose Transport and Spermidine Exporters
2.8. TMP Resistance Is Based on Reduced Cell Growth and Persister Formation
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Preparation of TCAT Solutions
4.3. Protein and DNA Sequences
4.4. Overexpression of E. coli DHFR (DHFR) Protein
4.5. Purification of DHFR Protein
4.6. E. coli DHFR Inhibition Assay
4.7. Minimal Inhibitory Concentration (MIC) Determination
4.8. Generation of Resistant Strains
4.9. Genomic DNA Isolation and Sequencing
4.10. Total RNA Isolation and Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Danner, M.C.; Robertson, A.; Behrends, V.; Reiss, J. Antibiotic pollution in surface fresh waters: Occurrence and effects. Sci. Total Environ. 2019, 664, 793–804. [Google Scholar] [CrossRef]
- Kramer, N.E.; Smid, E.J.; Kok, J.; Kruijff, B.; Kuipers, O.P.; Breukink, E. Resistance of Gram-positive bacteria to nisin is not determined by Lipid II levels. FEMS Microbiol. Lett. 2004, 239, 157–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundqvist, M. Reversibility of antibiotic resistance. Upsala J. Med. Sci. 2014, 119, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Casals-Pascual, C.; Vergara, A.; Vila, J. Intestinal microbiota and antibiotic resistance: Perspectives and solutions. Hum. Microbiome J. 2018, 9, 11–15. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerch, M.M.; Hansen, M.J.; van Dam, G.M.; Szymanski, W.; Feringa, B.L. Emerging targets in photopharmacology. Angew. Chem. Int. Ed. 2016, 55, 10978–10999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hüll, K.; Morstein, J.; Trauner, D. In vivo photopharmacology. Chem. Rev. 2018, 118, 10710–10747. [Google Scholar] [CrossRef]
- Hoorens, M.W.H.; Szymanski, W. Reversible, spatial and temporal control over protein activity using light. Trends Biochem. Sci. 2018, 43, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Welleman, I.M.; Hoorens, M.W.H.; Feringa, B.L.; Boersma, H.H.; Szymański, W. Photoresponsive molecular tools for emerging applications of light in medicine. Chem. Sci. 2020, 11, 11672–11691. [Google Scholar] [CrossRef]
- Velema, W.A.; Van Der Berg, J.P.; Hansen, M.J.; Szymanski, W.; Driessen, A.J.M.; Feringa, B.L. Optical control of antibacterial activity. Nat. Chem. 2013, 5, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Hansen, M.J.; Driessen, A.J.M.; Szymanski, W.; Feringa, B.L. Photocontrol of antibacterial activity: Shifting from UV to red light activation. J. Am. Chem. Soc. 2017, 139, 17979–17986. [Google Scholar] [CrossRef]
- Velema, W.A.; Hansen, M.J.; Lerch, M.M.; Driessen, A.J.M.; Szymanski, W.; Feringa, B.L. Ciprofloxacin-Photoswitch Conjugates: A facile strategy for photopharmacology. Bioconjug. Chem. 2015, 26, 2592–2597. [Google Scholar] [CrossRef] [PubMed]
- Babii, O.; Afonin, S.; Berditsch, M.; Reiβer, S.; Mykhailiuk, P.K.; Kubyshkin, V.S.; Steinbrecher, T.; Ulrich, A.S.; Komarov, I.V. Controlling biological activity with light: Diarylethene-containing cyclic peptidomimetics. Angew. Chem. Int. Ed. 2014, 53, 3392–3395. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Li, M.; Zhang, H.; Guo, H.; Ya, H.; Yin, J. Synthesis and properties of dithienylethene-functionalized switchable antibacterial agents. Org. Biomol. Chem. 2018, 16, 6988–6997. [Google Scholar] [CrossRef]
- Beharry, A.A.; Woolley, G.A. Azobenzene photoswitches for biomolecules. Chem. Soc. Rev. 2011, 40, 4422–4437. [Google Scholar] [CrossRef]
- Dong, M.; Babalhavaeji, A.; Samanta, S.; Beharry, A.A.; Woolley, G.A. Red-shifting azobenzene photoswitches for in vivo use. Acc. Chem. Res. 2015, 48, 2662–2670. [Google Scholar] [CrossRef]
- Weissleder, R. A clearer vision for in vivo imaging: Progress continues in the development of smaller, more penetrable probes for biological imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef]
- Huovinen, P. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 2001, 32, 1608–1614. [Google Scholar] [CrossRef] [Green Version]
- Gleckman, R.; Blagg, N.; Joubert, D.W. Trimethoprim: Mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharm. J. Hum. Pharm. Drug Ther. 1981, 1, 14–19. [Google Scholar] [CrossRef] [PubMed]
- British Medical Journal. Bacterial resistance to trimethoprim. Br. Med. J. 1980. [Google Scholar] [CrossRef] [Green Version]
- Brolund, A.; Sundqvist, M.; Kahlmeter, G.; Grape, M. Molecular characterisation of trimethoprim resistance in Escherichia coli and klebsiella pneumoniae during a two year intervention on trimethoprim use. PLoS ONE 2010, 5, e9233. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hossein Khademi, S.M.; Sazinas, P.; Jelsbak, L. Within-host adaptation mediated by intergenic evolution in pseudomonas aeruginosa. Genome Biol. Evol. 2019, 11, 1385–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, E.A.; Graves, J.F.; Hite, L.A.; Parker, C.T.; Schnaitman, C.A. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli k-12: Insertion mutagenesis of the rfa locus. J. Bacteriol. 1990, 172, 12–25. [Google Scholar] [CrossRef] [Green Version]
- Zgurskaya, H.I.; Krishnamoorthy, G.; Ntreh, A.; Lu, S. Mechanism and function of the outer membrane channel TolC in multidrug resistance and physiology of enterobacteria. Front. Microbiol. 2011, 2, 189. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, A.; van der Meulen, S.; Kuipers, O.P.; Kok, J. T-REx: Transcriptome analysis webserver for RNA-seq Expression data. BMC Genom. 2015, 16, 663. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.; Casjens, S.; Krishnaswamy, S. Analysis of the lambdoid prophage element e14 in the E. coli K-12 genome. BMC Microbiol. 2004, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Christensen-Dalsgaard, M.; Jørgensen, M.G.; Gerdes, K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010, 75, 333–348. [Google Scholar] [CrossRef] [Green Version]
- Woodgate, R.; Ennis, D.G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. MGG Mol. Gen. Genet. 1991, 229, 10–16. [Google Scholar] [CrossRef]
- Méjean, V.; Lobbi-Nivol, C.; Lepelletier, M.; Giordano, G.; Chippaux, M.; Pascal, M.-C. TMAO anaerobic respiration in Escherichia coli: Involvement of the tor operon. Mol. Microbiol. 1994, 11, 1169–1179. [Google Scholar] [CrossRef]
- Reid, A.N.; Whitfield, C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: Evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 2005, 187, 5470–5481. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, G.; Andrianopoulos, K.; Hobbs, M.; Reeves, P.R. Organization of the Escherichia coli k-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1996, 178, 4885–4893. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Beis, K.; Nesper, J.; Brunkan-LaMontagne, A.L.; Clarke, B.R.; Whitfield, C.; Naismith, J.H. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 2006, 444, 226–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Biase, D.; Tramonti, A.; Bossa, F.; Visca, P. The response to stationary-phase stress conditions in Escherichia coli: Role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999, 32, 1198–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unoson, C.; Wagner, E.G.H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008, 70, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Bougdour, A.; Wickner, S.; Gottesman, S. Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev. 2006, 20, 884–897. [Google Scholar] [CrossRef] [Green Version]
- Mealman, T.D.; Blackburn, N.J.; McEvoy, M.M. Metal Export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. In Current Topics in Membranes; Academic Press Inc.: Cambridge, MA, USA, 2012; Volume 69, pp. 163–196. [Google Scholar]
- Faubladiert, M.; Bouche, J.-P. Division Inhibition Gene dicF of Escherichia coli Reveals a Widespread Group of Prophage Sequences in Bacterial Genomes. J. Bacteriol. 1994, 176, 1150–1156. [Google Scholar] [CrossRef] [Green Version]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef] [PubMed]
- Nagakubo, S.; Nishino, K.; Hirata, T.; Yamaguchi, A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 2002, 184, 4161–4167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meadows, J.A.; Wargo, M.J. Carnitine in bacterial physiology and metabolism. Microbiology 2015, 161, 1161–1174. [Google Scholar] [CrossRef]
- Dumont, E.; Vergalli, J.; Pajovic, J.; Bhamidimarri, S.P.; Morante, K.; Wang, J.; Lubriks, D.; Suna, E.; Stavenger, R.A.; Winterhalter, M.; et al. Mechanistic aspects of maltotriose-conjugate translocation to the Gram-negative bacteria cytoplasm. Life Sci. Alliance 2019, 2, e201800242. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.K.; Sternberg, N.L. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: Application to DMP 840, a tumoricidal agent. Proc. Natl. Acad. Sci. USA 1995, 92, 8950–8954. [Google Scholar] [CrossRef] [Green Version]
- Higashi, K.; Ishigure, H.; Demizu, R.; Uemura, T.; Nishino, K.; Yamaguchi, A.; Kashiwagi, K.; Igarashi, K. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008, 190, 872–878. [Google Scholar] [CrossRef] [Green Version]
- Peralta, D.R.; Adler, C.; Corbalán, N.S.; Paz García, E.C.; Pomares, M.F.; Vincent, P.A. Enterobactin as part of the oxidative stress response repertoire. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Guest, R.L.; Court, E.A.; Waldon, J.L.; Schock, K.A.; Raivio, T.L. Impaired efflux of the siderophore enterobactin induces envelope stress in Escherichia coli. Front. Microbiol. 2019, 10, 2776. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, A.D.; Deisenhofer, J. TonB-dependent receptors—Structural perspectives. Biochim. Biophys. Acta Biomembr. 2002, 1565, 318–332. [Google Scholar] [CrossRef] [Green Version]
- Griggs, D.W.; Tharp, B.B.; Konisky, J. Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J. Bacteriol. 1987, 169, 5343–5352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atlung, T.; Brondsted, L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J. Bacteriol. 1994, 176, 5414–5422. [Google Scholar] [CrossRef] [Green Version]
- Beauchene, N.A.; Mettert, E.L.; Moore, L.J.; Keleş, S.; Willey, E.R.; Kiley, P.J. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 12261–12266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keren, I.; Shah, D.; Spoering, A.; Kaldalu, N.; Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004, 186, 8172–8180. [Google Scholar] [CrossRef] [Green Version]
- Griffith, J.K.; Baker, M.E.; Rouch, D.A.; Page, M.G.P.; Skurray, R.A.; Paulsen, I.T.; Chater, K.F.; Baldwin, S.A.; Henderson, P.J.F. Membrane transport proteins: Implications of sequence comparisons. Curr. Opin. Cell Biol. 1992, 4, 684–695. [Google Scholar] [CrossRef]
- Nishino, K.; Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Weber, K.D.; Qiu, Y.; Kiley, P.J.; Blattner, F.R. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 2005, 187, 1135–1160. [Google Scholar] [CrossRef] [Green Version]
- Kaczanowska, M.; Rydén-Aulin, M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef] [Green Version]
- Svenningsen, M.S.; Veress, A.; Harms, A.; Mitarai, N.; Semsey, S. Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta Biomembr. 2005, 1717, 67–88. [Google Scholar] [CrossRef] [Green Version]
- Schober, A.F.; Mathis, A.D.; Ingle, C.; Park, J.O.; Chen, L.; Rabinowitz, J.D.; Junier, I.; Rivoire, O.; Reynolds, K.A. A Two-enzyme adaptive unit within bacterial folate metabolism. Cell Rep. 2019, 27, 3359–3370. [Google Scholar] [CrossRef] [PubMed]
- Way, J.C.; Davis, M.A.; Morisato, D.; Roberts, D.E.; Kleckner, N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 1984, 32, 369–379. [Google Scholar] [CrossRef]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutiérrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; van den Berg, B.; Winterhalter, M.; et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Iwakura, M.; Jones, B.E.; Luo, J.; Matthews, R. A strategy for testing the suitability of cysteine replacements in dihydrofolate reductase from Escherichia coli. J. Biochem. 1995, 117, 480–488. [Google Scholar] [CrossRef]
- Patel, J.B.; Cockerill, R.F.; Bradford, P.A.; Eliopoulos, G.M.; Hindler, J.A.; Jenkins, S.G.; Lewis II, J.S.; Limbago, B.; Miller, L.A.; Nicolau, D.P.; et al. CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; The Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; ISBN 1-56238-988-2. [Google Scholar]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef]

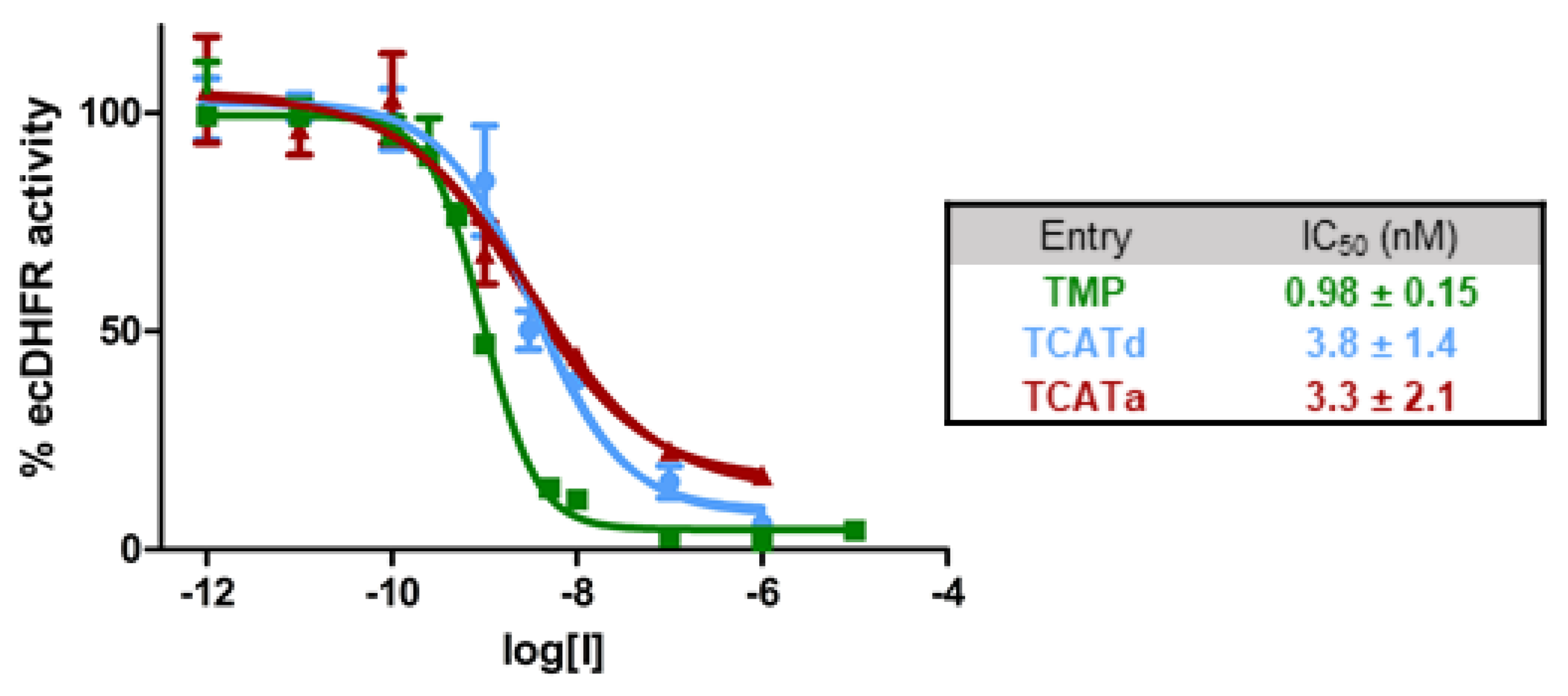


| Strain | MIC TMP (µM) | MIC TCATa (µM) | MIC TCATd (µM) | Number of Generations |
|---|---|---|---|---|
| E. coli CS1562 | 0.05 | 4.9 | 80.1 | 32 |
| TMPR | 1.4 | 21 | ||
| TCATaR | 49.3 | 23 | ||
| TCATdR | 98.5 | 12 |
| Predicted Mutations | ||||||
|---|---|---|---|---|---|---|
| Mutation | Annotation | Gene | Description | Evidence | Seq Id | Position |
| G→A | Intergenic (+202/−293) | aqpZ→/→lysO | aquaporin Z/L lysine exporter LysO | RA | NODE_2 | 134,755 |
| Δ2bp | Intergenic (+2/−2) | Ecoli_02842→/→Ecoli_02843 | tRNA Val/tRNA Lys | RA | NODE_20 | 45,821 |
| Δ1bp | Intergenic (+3/−2) | Ecoli_02844→/→Ecoli_02845 | tRNA Val/tRNA Lys | RA | NODE_20 | 46,127 |
| A→G | Intergenic (+1023/+707) | chaA→/←kdsA | sodium potassium/proton antiporter ChaA/3 deoxy 8 phosphooctulonate synthase | RA | NODE_4 | 17,237 |
| A→C | Intergenic (+1387/+343) | chaA→/←kdsA | sodium potassium/proton antiporter ChaA/3 deoxy 8 phosphooctulonate synthase | RA | NODE_4 | 17,601 |
| Δ1bp | Intergenic (+1420/+310) | chaA→/←kdsA | sodium potassium/proton antiporter ChaA/3 deoxy 8 phosphooctulonate synthase | RA | NODE_4 | 17,634 |
| +A | Intergenic (+1425/+305) | chaA→/←kdsA | sodium potassium/proton antiporter ChaA/3 deoxy 8 phosphooctulonate synthase | RA | NODE_4 | 17,639 |
| C→A | Intergenic (+1428/+302) | chaA→/←kdsA | sodium potassium/proton antiporter ChaA/3 deoxy 8 phosphooctulonate synthase | RA | NODE_4 | 17,642 |
| T→C | Intergenic (−/−) | −/− | −/− | RA | NODE_97 | 18 |
| T→G | Intergenic (−/−) | −/− | −/− | RA | NODE_97 | 65 |
| G→A | Intergenic (−/−) | −/− | −/− | RA | NODE_97 | 98 |
| C→T | Intergenic (−/−) | −/− | −/− | RA | NODE_97 | 104 |
| G→T | L210L (CTG→CTT) | Ecoli_03722 → | IS5 like element ISKpn26 family transposase | RA | NODE_39 | 4254 |
| Predicted Mutations | ||||||
|---|---|---|---|---|---|---|
| Mutation | Annotation | Gene | Description | Evidence | Seq Id | Position |
| Δ1bp | coding (87/1257 nt) | Ecoli_01894 → | MFS transporter | RA | NODE_10 | 108,811 |
| Δ2934 bp | Ecoli_03163 [tolC_2] | Ecoli_03163, tetR(A), tet(A), tetC, [tolC_2] | MC, JC → | NODE_26 | 7555 | |
| (A) Factors | (B) Contrasts | |
|---|---|---|
| Experiment | Strain | |
| C | E. coli CS1562 (propagated parent strain as a control) | |
| TCATaR | TCATa resistant isolate | TCATaR-C |
| TCATdR | TCATd resistant isolate | TCATdR-C |
| TMPR | TMP- resistant isolate | TMPR-C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauxen, A.I.; Kobauri, P.; Wegener, M.; Hansen, M.J.; Galenkamp, N.S.; Maglia, G.; Szymanski, W.; Feringa, B.L.; Kuipers, O.P. Mechanism of Resistance Development in E. coli against TCAT, a Trimethoprim-Based Photoswitchable Antibiotic. Pharmaceuticals 2021, 14, 392. https://doi.org/10.3390/ph14050392
Lauxen AI, Kobauri P, Wegener M, Hansen MJ, Galenkamp NS, Maglia G, Szymanski W, Feringa BL, Kuipers OP. Mechanism of Resistance Development in E. coli against TCAT, a Trimethoprim-Based Photoswitchable Antibiotic. Pharmaceuticals. 2021; 14(5):392. https://doi.org/10.3390/ph14050392
Chicago/Turabian StyleLauxen, Anna I., Piermichele Kobauri, Michael Wegener, Mickel J. Hansen, Nicole S. Galenkamp, Giovanni Maglia, Wiktor Szymanski, Ben L. Feringa, and Oscar P. Kuipers. 2021. "Mechanism of Resistance Development in E. coli against TCAT, a Trimethoprim-Based Photoswitchable Antibiotic" Pharmaceuticals 14, no. 5: 392. https://doi.org/10.3390/ph14050392
APA StyleLauxen, A. I., Kobauri, P., Wegener, M., Hansen, M. J., Galenkamp, N. S., Maglia, G., Szymanski, W., Feringa, B. L., & Kuipers, O. P. (2021). Mechanism of Resistance Development in E. coli against TCAT, a Trimethoprim-Based Photoswitchable Antibiotic. Pharmaceuticals, 14(5), 392. https://doi.org/10.3390/ph14050392







