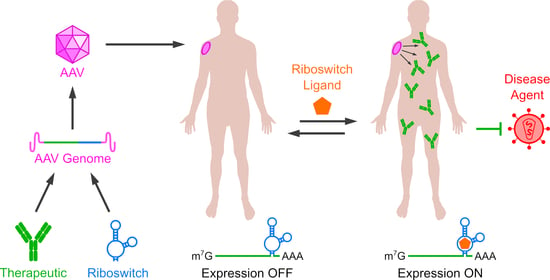
Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors
Abstract
:1. Introduction
2. Riboswitch Regulation of Transgene Expression in Mammals
2.1. Riboswitches Regulating mRNA Processing
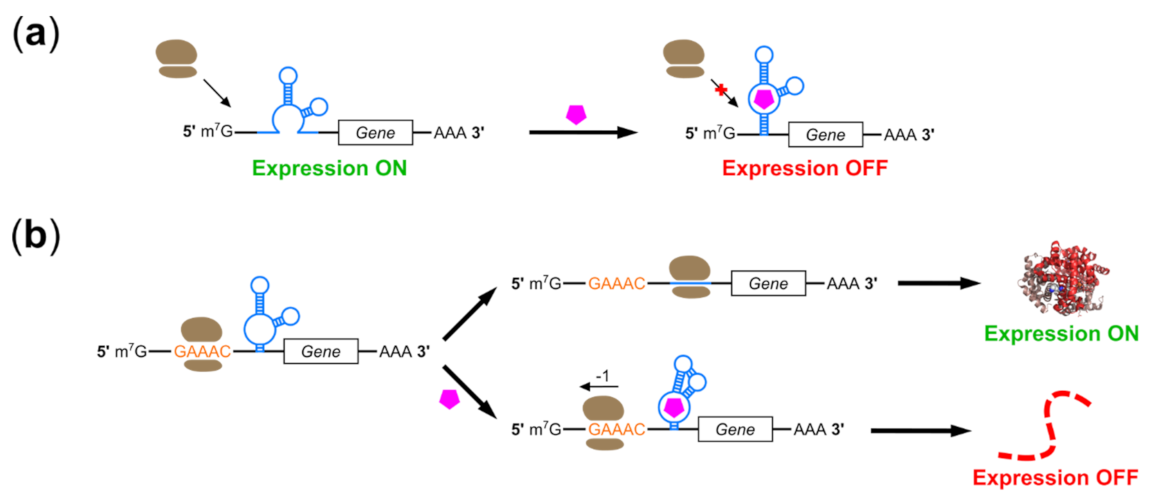
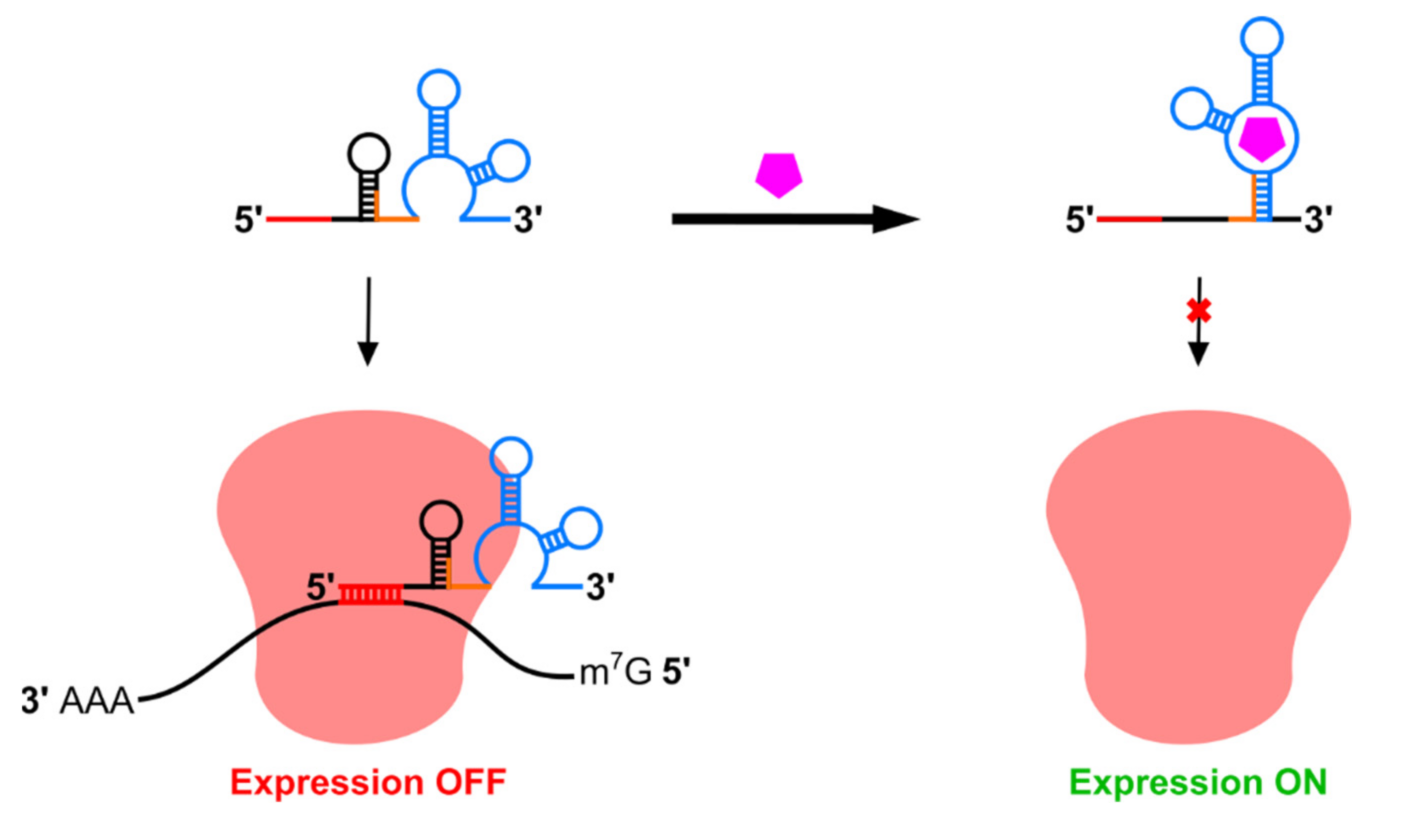
2.2. Riboswitches Controlling Translation Initiation
2.3. Riboswitches Controlling Alternative Initiation Mechanisms
2.4. Programmed Ribosomal Frameshifting Switches
2.5. RNA Interference-Based Riboswitches
2.6. Catalytic Riboswitches
2.7. Improving the Function of Aptazyme Riboswitches
2.8. Regulation of CRISPR-Cas Activity by Riboswitches
| Switch Construct | Mechanism | Polarity | Ligand | Fold Regulation | Reference |
|---|---|---|---|---|---|
| A2 | Polyadenylation | Off | Guanine | 5.2 | Spöring et al. 2020 [66] |
| L2 | Splicing | On | Tetracycline | 5.7 | Vogel et al. 2018 [70] |
| Tet13_el | Splicing | On | Tetracycline | 16.9 | Finke et al. 2021 [71] |
| AS325 W-P9 6T8T | Trans-splicing | On | Theophylline | 2 | Kim et al. 2014 [75] |
| SVH2βgal | Translation roadblock | Off | Hoechst dyes | 10 | Werstuck and Green 1998 [79] |
| CFS-RX GAAA | Translation roadblock | Off | Ciprofloxacin | 1.8 | Groher et al. 2018 [85] |
| R22 | Translation roadblock | Off | Theophylline | 10 | Liu et al. 2018 [89] |
| R26 | Translation roadblock | Off | Tetracycline | 10 | Liu et al. 2018 [89] |
| 68 metH | Programmed ribosomal frameshifting | On/Off | Adenosine-2’,3’-dialdehyde | 4.39 | Chou et al. 2010 [105] |
| Theo-OFF2-MMTV | Programmed ribosomal frameshifting | On/Off | Theophylline | 6 | Hsu et al. 2014 [108] |
| TheoOFF2 SARS | Programmed ribosomal frameshifting | On/Off | Theophylline | 1.5 | Lin and Chang 2016 [109] |
| M1-VPK | Programmed ribosomal frameshifting | On/Off | NCT8 | 9.1 | Matsumoto et al. 2018 [110] |
| pE19T | miRNA processing | On | Theophylline | 3 | An et al. 2006 [118] |
| th1 | miRNA processing | On | Theophylline | 4.1 | Beisel et al. 2011 [120] |
| pRzTheo-miREGFP | miRNA processing | Off | Theophylline | 4 | Kumar et al. 2009 [122] |
| β(1x) | miRNA processing | On | Folinic acid | 3.5 | Wong et al. 2018 [123] |
| miR-378a-CT | miRNA processing | On | Theophylline | 5.7 | Pollak et al. 2021 [124] |
| tac210 | miRNA site accessibility | On | Tetracycline | 19 | Mou et al. 2018 [125] |
| P1-F5, 5.3 | mRNA self-cleavage | Off | Theophylline | 6 | Ausländer et al. 2010 [130] |
| 7c4x | mRNA self-cleavage | On | Guanine | 6.7 | Mustafina et al. 2020 [133] |
| K19 | mRNA self-cleavage | On | Tetracycline | 8.7 | Beilstein et al. 2015 [136] |
| GuaM8HDV | mRNA self-cleavage | Off | Guanine | 29.5 | Nomura et al. 2013 [140] |
| Tc40 | mRNA self-cleavage | Off | Tetracycline | 24 | Zhong et al. 2016 [153] |
| Gua_K3 | mRNA self-cleavage | On | Guanine | 4 | Stifel et al. 2019 [168] |
| Theo-HHR-B | mRNA self-cleavage | Off | Theophylline | 4.8 | Zhang et al. 2017 [169] |
| TAP1 | mRNA self-cleavage | Off | Theophylline | 7 | Pu et al. 2020 [170] |
| XanACGAG | mRNA self-cleavage | On | Hypoxanthine | 6.8 | Xiang et al. 2019 [171] |
| FolUGAAG | mRNA self-cleavage | On | (6R,S)-folinic acid | 5.3 | Xiang et al. 2019 [171] |
| cdG-CGUAA | mRNA self-cleavage | On | Cyclic di-GMP | 2 | Xiang et al. 2019 [171] |
| sgRNA4 | CRISPR-Cas gRNA accessibility | Off | Tetracycline | 4.25 | Liu et al. 2016 [187] |
| Theophylline-agRNA | CRISPR-Cas gRNA cleavage | On/Off | Theophylline | 3 | Tang et al. 2017 [188] |
| Guanine-agRNA | CRISPR-Cas gRNA cleavage | On/Off | Guanine | 4.8 | Tang et al. 2017 [188] |
2.9. Deoxyribozyme Switches
3. Therapeutic Applications of Riboswitches
3.1. Regulation of Erythropoeitin Expression
3.2. Regulation of Vectored Immunoprophylaxis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buschmann, M.; Carrasco, M.; Alishetty, S.; Paige, M.; Alameh, M.; Weissman, D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines 2021, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Hastie, E.; Samulski, R.J. Adeno-Associated Virus at 50: A Golden Anniversary of Discovery, Research, and Gene Therapy Success—A Personal Perspective. Hum. Gene Ther. 2015, 26, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Atchison, R.W.; Casto, B.C.; Hammon, W.M. Adenovirus-Associated Defective Virus Particles. Science 1965, 149, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020, 28, 723–746. [Google Scholar] [CrossRef] [PubMed]
- Deyle, D.R.; Russell, D.W. Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 2009, 11, 442–447. [Google Scholar] [PubMed]
- Hirata, R.; Chamberlain, J.; Dong, R.; Russell, D.W. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002, 20, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Asokan, A.; Samulski, R.J. Adeno-associated Virus Serotypes: Vector Toolkit for Human Gene Therapy. Mol. Ther. 2006, 14, 316–327. [Google Scholar] [CrossRef]
- Davidsson, M.; Wang, G.; Aldrin-Kirk, P.; Cardoso, T.; Nolbrant, S.; Hartnor, M.; Mudannayake, J.; Parmar, M.; Björklund, T. A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism. Proc. Natl. Acad. Sci. USA 2019, 116, 27053–27062. [Google Scholar] [CrossRef] [Green Version]
- Jackson, C.B.; Richard, A.S.; Ojha, A.; Conkright, K.A.; Trimarchi, J.M.; Bailey, C.C.; Alpert, M.D.; Kay, M.A.; Farzan, M.; Choe, H. AAV vectors engineered to target insulin receptor greatly enhance intramuscular gene delivery. Mol. Ther. Methods Clin. Dev. 2020, 19, 496–506. [Google Scholar] [CrossRef]
- Mccarty, D.M.E.; Monahan, P.; Samulski, R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001, 8, 1248–1254. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhao, W.; Zhong, L.; Han, Z.; Li, B.; Ma, W.; Weigel-Kelley, K.A.; Warrington, K.H.; Srivastava, A. Self-Complementary Recombinant Adeno-Associated Viral Vectors: Packaging Capacity and The Role of Rep Proteins in Vector Purity. Hum. Gene Ther. 2007, 18, 171–182. [Google Scholar] [CrossRef]
- Trapani, I. Adeno-Associated Viral Vectors as a Tool for Large Gene Delivery to the Retina. Genes 2019, 10, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.-H.; Yu, N.-K.; Baek, G.-C.; Bakes, J.; Seo, D.; Nam, H.J.; Baek, S.H.; Lim, C.-S.; Lee, Y.-S.; Kaang, B.-K. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain 2014, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Penaud-Budloo, M.; Le Guiner, C.; Nowrouzi, A.; Toromanoff, A.; Chérel, Y.; Chenuaud, P.; Schmidt, M.; von Kalle, C.; Rolling, F.; Moullier, P.; et al. Adeno-Associated Virus Vector Genomes Persist as Episomal Chromatin in Primate Muscle. J. Virol. 2008, 82, 7875–7885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeler, A.M.; Flotte, T.R. Recombinant Adeno-Associated Virus Gene Therapy in Light of Luxturna (and Zolgensma and Glybera): Where Are We, and How Did We Get Here? Annu. Rev. Virol. 2019, 6, 601–621. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef]
- Li, T.; Chen, X.; Qian, Y.; Shao, J.; Li, X.; Liu, S.; Zhu, L.; Zhao, Y.; Ye, H.; Yang, Y. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice. Nat. Commun. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-On Systems for Doxycycline-inducible Gene Expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef] [Green Version]
- Moncion, A.; Harmon, J.; Li, Y.; Natla, S.; Farrell, E.C.; Kripfgans, O.D.; Stegemann, J.P.; Martín-Saavedra, F.M.; Vilaboa, N.; Franceschi, R.T.; et al. Spatiotemporally-controlled transgene expression in hydroxyapatite-fibrin composite scaffolds using high intensity focused ultrasound. Biomaterials 2019, 194, 14–24. [Google Scholar] [CrossRef]
- Kallunki, T.; Barisic, M.; Jäättelä, M.; Liu, B. How to Choose the Right Inducible Gene Expression System for Mammalian Studies? Cells 2019, 8, 796. [Google Scholar] [CrossRef] [Green Version]
- Strobel, B.; Klauser, B.; Hartig, J.S.; Lamla, T.; Gantner, F.; Kreuz, S. Riboswitch-mediated Attenuation of Transgene Cytotoxicity Increases Adeno-associated Virus Vector Yields in HEK-293 Cells. Mol. Ther. 2015, 23, 1582–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, R.W. Complexity of immune responses to AAV transgene products—Example of factor IX. Cell. Immunol. 2019, 342, 103658. [Google Scholar] [CrossRef] [PubMed]
- Yokobayashi, Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells. Curr. Opin. Chem. Biol. 2019, 52, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Hirao, I.; Ellington, A.D. Re-creating the RNA world. Curr. Biol. 1995, 5, 1017–1022. [Google Scholar] [CrossRef] [Green Version]
- Breaker, R.R. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2010, 4, a003566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Breaker, R. Rational design of allosteric ribozymes. Chem. Biol. 1997, 4, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Groher, F.; Suess, B. Synthetic riboswitches—A tool comes of age. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1839, 964–973. [Google Scholar] [CrossRef]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic Control by a Metabolite Binding mRNA. Chem. Biol. 2002, 9, 1043–1049. [Google Scholar] [CrossRef] [Green Version]
- Winkler, W.C.; Nahvi, A.; Breaker, R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nat. Cell Biol. 2002, 419, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Cohen-Chalamish, S.; Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 2002, 99, 15908–15913. [Google Scholar] [CrossRef] [Green Version]
- Breaker, R.R. Riboswitches and Translation Control. Cold Spring Harb. Perspect. Biol. 2018, 10, a032797. [Google Scholar] [CrossRef]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef]
- Chahal, J.; Gebert, L.; Gan, H.H.; Camacho, E.; Gunsalus, K.C.; Macrae, I.J.; Sagan, S.M. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5′ terminus. Nucleic Acids Res. 2019, 47, 5307–5324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbaiah, K.C.V.; Hedaya, O.; Wu, J.; Jiang, F.; Yao, P. Mammalian RNA switches: Molecular rheostats in gene regulation, disease, and medicine. Comput. Struct. Biotechnol. J. 2019, 17, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Arachchilage, G.M.; Atilho, R.; Stav, S.; Higgs, G.; Breaker, R. Genome-wide Discovery of Rare Riboswitches in Bacteria. FASEB J. 2019, 33, 778. [Google Scholar] [CrossRef]
- Borujeni, A.E.; Mishler, D.M.; Wang, J.; Huso, W.; Salis, H.M. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2015, 44, 1–13. [Google Scholar] [CrossRef]
- Soukup, G.A.; Breaker, R.R. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. USA 1999, 96, 3584–3589. [Google Scholar] [CrossRef] [Green Version]
- Haines, M.C.; Storch, M.; Oyarzún, D.A.; Stan, G.-B.; Baldwin, G.S. Riboswitch identification using Ligase-Assisted Selection for the Enrichment of Responsive Ribozymes (LigASERR). Synth. Biol. 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Martini, L.; Ellington, A.D.; Mansy, S.S. An in vitro selection for small molecule induced switching RNA molecules. Methods 2016, 106, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Davidson, M.E.; Harbaugh, S.V.; Chushak, Y.G.; Stone, M.O.; Kelley-Loughnane, N. Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli Cells. ACS Chem. Biol. 2013, 8, 234–241. [Google Scholar] [CrossRef]
- Fowler, C.C.; Brown, E.D.; Li, Y. A FACS-Based Approach to Engineering Artificial Riboswitches. ChemBioChem 2008, 9, 1906–1911. [Google Scholar] [CrossRef]
- Lynch, S.A.; Desai, S.K.; Sajja, H.K.; Gallivan, J.P. A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function. Chem. Biol. 2007, 14, 173–184. [Google Scholar] [CrossRef] [Green Version]
- Dixon, N.; Duncan, J.N.; Geerlings, T.; Dunstan, M.S.; McCarthy, J.E.G.; Leys, D.; Micklefield, J. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. USA 2010, 107, 2830–2835. [Google Scholar] [CrossRef] [Green Version]
- Nomura, Y.; Yokobayashi, Y. Reengineering a Natural Riboswitch by Dual Genetic Selection. J. Am. Chem. Soc. 2007, 129, 13814–13815. [Google Scholar] [CrossRef]
- Townshend, B.; Kennedy, A.B.; Xiang, J.S.; Smolke, C.D. High-throughput cellular RNA device engineering. Nat. Methods 2015, 12, 989–994. [Google Scholar] [CrossRef]
- Stoddard, C.D.; Widmann, J.; Trausch, J.J.; Marcano-Velázquez, J.G.; Knight, R.; Batey, R.T. Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch. J. Mol. Biol. 2013, 425, 1596–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigand, J.; Gottstein-Schmidtke, S.R.; Demolli, S.; Groher, F.; Duchardt-Ferner, E.; Wöhnert, J.; Suess, B. Sequence Elements Distal to the Ligand Binding Pocket Modulate the Efficiency of a Synthetic Riboswitch. ChemBioChem 2014, 15, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Topp, S.; Reynoso, C.M.K.; Seeliger, J.C.; Goldlust, I.S.; Desai, S.K.; Murat, D.; Shen, A.; Puri, A.W.; Komeili, A.; Bertozzi, C.R.; et al. Synthetic Riboswitches That Induce Gene Expression in Diverse Bacterial Species. Appl. Environ. Microbiol. 2010, 76, 7881–7884. [Google Scholar] [CrossRef] [Green Version]
- Kent, R.; Dixon, N. Systematic Evaluation of Genetic and Environmental Factors Affecting Performance of Translational Riboswitches. ACS Synth. Biol. 2019, 8, 884–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceres, P.; Garst, A.D.; Marcano-Velázquez, J.G.; Batey, R.T. Modularity of Select Riboswitch Expression Platforms Enables Facile Engineering of Novel Genetic Regulatory Devices. ACS Synth. Biol. 2013, 2, 463–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallberg, Z.F.; Su, Y.; Kitto, R.Z.; Hammond, M.C. Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem. 2017, 86, 515–539. [Google Scholar] [CrossRef]
- Leamy, K.A.; Assmann, S.M.; Mathews, D.H.; Bevilacqua, P.C. Bridging the gap betweenin vitroandin vivoRNA folding. Q. Rev. Biophys. 2016, 49, e10. [Google Scholar] [CrossRef] [Green Version]
- Yamagami, R.; Huang, R.; Bevilacqua, P.C. Cellular Concentrations of Nucleotide Diphosphate-Chelated Magnesium Ions Accelerate Catalysis by RNA and DNA Enzymes. Biochemistry 2019, 58, 3971–3979. [Google Scholar] [CrossRef]
- Etzel, M.; Mörl, M. Synthetic Riboswitches: From Plug and Pray toward Plug and Play. Biochemistry 2017, 56, 1181–1198. [Google Scholar] [CrossRef]
- Corbett, A.H. Post-transcriptional regulation of gene expression and human disease. Curr. Opin. Cell Biol. 2018, 52, 96–104. [Google Scholar] [CrossRef]
- Assche, E.E.; Puyvelde, S.E.; Evanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef]
- Wachsmuth, M.; Findeiss, S.; Weissheimer, N.; Stadler, P.F.; Mörl, M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 2012, 41, 2541–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceres, P.; Trausch, J.J.; Batey, R.T. Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Res. 2013, 41, 10449–10461. [Google Scholar] [CrossRef]
- Domin, G.; Findeiß, S.; Wachsmuth, M.; Will, S.; Stadler, P.F.; Mörl, M. Applicability of a computational design approach for synthetic riboswitches. Nucleic Acids Res. 2016, 45, 4108–4119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scull, C.E.; Dandpat, S.S.; Romero, R.A.; Walter, N.G. Transcriptional Riboswitches Integrate Timescales for Bacterial Gene Expression Control. Front. Mol. Biosci. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Braselmann, E.; Wierzba, A.J.; Polaski, J.T.; Chromiński, M.; Holmes, Z.E.; Hung, S.-T.; Batan, D.; Wheeler, J.R.; Parker, R.; Jimenez, R.; et al. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol. 2018, 14, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Spöring, M.; Boneberg, R.; Hartig, J.S. Aptamer-Mediated Control of Polyadenylation for Gene Expression Regulation in Mammalian Cells. ACS Synth. Biol. 2020, 9, 3008–3018. [Google Scholar] [CrossRef]
- Li, S.; Breaker, R.R. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res. 2013, 41, 3022–3031. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-S.; Gusti, V.; Dery, K.J.; Gaur, R.K. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 2008, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Weigand, J.E.; Suess, B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007, 35, 4179–4185. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.; Weigand, J.E.; Kluge, B.; Grez, M.; Suess, B. A small, portable RNA device for the control of exon skipping in mammalian cells. Nucleic Acids Res. 2018, 46, e48. [Google Scholar] [CrossRef] [Green Version]
- Finke, M.; Brecht, D.; Stifel, J.; Gense, K.; Gamerdinger, M.; Hartig, J.S. Efficient splicing-based RNA regulators for tetracycline-inducible gene expression in human cell culture and C. elegans. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef]
- Culler, S.J.; Hoff, K.G.; Smolke, C.D. Reprogramming Cellular Behavior with RNA Controllers Responsive to Endogenous Proteins. Science 2010, 330, 1251–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mol, A.A.; Groher, F.; Schreiber, B.; Rühmkorff, C.; Suess, B. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module. Nucleic Acids Res. 2019, 47, e132. [Google Scholar] [CrossRef]
- Mol, A.A.; Vogel, M.; Suess, B. Inducible nuclear import by TetR aptamer-controlled 3′ splice site selection. RNA 2021, 27, 234–241. [Google Scholar] [CrossRef]
- Kim, J.; Jeong, S.; Kertsburg, A.; Soukup, G.A.; Lee, S.-W. Conditional and Target-Specific Transgene Induction through RNA Replacement Using an Allosteric Trans-Splicing Ribozyme. ACS Chem. Biol. 2014, 9, 2491–2495. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.-S.; Jung, H.-S.; Song, M.-S.; Cho, K.S.; Kim, S.-C.; Kimm, K.; Jeong, J.S.; Kim, I.-H.; Lee, S.-W. Specific Regression of Human Cancer Cells by Ribozyme-Mediated Targeted Replacement of Tumor-Specific Transcript. Mol. Ther. 2005, 12, 824–834. [Google Scholar] [CrossRef]
- Hong, S.-H.; Jeong, J.-S.; Lee, Y.-J.; Jung, H.-I.; Cho, K.-S.; Kim, C.-M.; Kwon, B.-S.; Sullenger, B.A.; Lee, S.-W.; Kim, I.-H. In Vivo Reprogramming of hTERT by Trans-splicing Ribozyme to Target Tumor Cells. Mol. Ther. 2008, 16, 74–80. [Google Scholar] [CrossRef]
- Ogawa, A. Rational design of artificial riboswitches based on ligand-dependent modulation of internal ribosome entry in wheat germ extract and their applications as label-free biosensors. RNA 2011, 17, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Werstuck, G. Controlling Gene Expression in Living Cells through Small Molecule-RNA Interactions. Science 1998, 282, 296–298. [Google Scholar] [CrossRef] [Green Version]
- Hanson, S.; Berthelot, K.; Fink, B.; McCarthy, J.E.G.; Suess, B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003, 49, 1627–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, I.; Garneau, P.; Pelletier, J. Inhibition of translation by RNA–small molecule interactions. RNA 2002, 8, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Grate, D.; Wilson, C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer–ligand complex. Bioorg. Med. Chem. 2001, 9, 2565–2570. [Google Scholar] [CrossRef]
- Kozak, M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 1986, 83, 2850–2854. [Google Scholar] [CrossRef] [Green Version]
- Babendure, J.R.; Ding, J.-H.; Tsien, R.Y. Control of mammalian translation by mRNA structure near caps. RNA 2006, 12, 851–861. [Google Scholar] [CrossRef] [Green Version]
- Groher, F.; Bofill-Bosch, C.; Schneider, C.; Braun, J.; Jager, S.; Geißler, K.; Hamacher, K.; Suess, B. Riboswitching with ciprofloxacin—development and characterization of a novel RNA regulator. Nucleic Acids Res. 2018, 46, 2121–2132. [Google Scholar] [CrossRef]
- Boussebayle, A.; Torka, D.; Ollivaud, S.; Braun, J.; Bofill-Bosch, C.; Dombrowski, M.; Groher, F.; Hamacher, K.; Suess, B. Next-level riboswitch development—Implementation of Capture-SELEX facilitates identification of a new synthetic riboswitch. Nucleic Acids Res. 2019, 47, 4883–4895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldfless, S.J.; Belmont, B.J.; De Paz, A.M.; Liu, J.F.; Niles, J.C. Direct and specific chemical control of eukaryotic translation with a synthetic RNA–protein interaction. Nucleic Acids Res. 2012, 40, e64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, T.S.; Smolke, C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005, 23, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J.; Chen, Z.; Huang, W.; Cai, Z. Synthesizing artificial devices that redirect cellular information at will. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakawa, S.; Oguro, A.; Ohtsu, T.; Imataka, H.; Sonenberg, N.; Nakamura, Y. RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA 2006, 12, 1825–1834. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, A. Rational construction of eukaryotic OFF-riboswitches that downregulate internal ribosome entry site-mediated translation in response to their ligands. Bioorg. Med. Chem. Lett. 2012, 22, 1639–1642. [Google Scholar] [CrossRef]
- Ogawa, A.; Masuoka, H.; Ota, T. Artificial OFF-Riboswitches That Downregulate Internal Ribosome Entry without Hybridization Switches in a Eukaryotic Cell-Free Translation System. ACS Synth. Biol. 2017, 6, 1656–1662. [Google Scholar] [CrossRef]
- Ogawa, A.; Itoh, Y. In Vitro Selection of RNA Aptamers Binding to Nanosized DNA for Constructing Artificial Riboswitches. ACS Synth. Biol. 2020, 9, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Murashige, Y.; Tabuchi, J.; Omatsu, T. Ligand-responsive upregulation of 3′ CITE-mediated translation in a wheat germ cell-free expression system. Mol. BioSyst. 2016, 13, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud-Gabardos, E.; Hantelys, F.; Morfoisse, F.; Chaufour, X.; Garmy-Susini, B.; Prats, A.-C. Internal ribosome entry site-based vectors for combined gene therapy. World J. Exp. Med. 2015, 5, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A. Ligand-Dependent Upregulation of Ribosomal Shunting. ChemBioChem 2013, 14, 1539–1543. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A. Engineering of Ribosomal Shunt-Modulating Eukaryotic ON Riboswitches by Using a Cell-Free Translation System. Methods Enzymol. 2015, 550, 109–128. [Google Scholar] [CrossRef]
- Yueh, A.; Schneider, R.J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996, 10, 1557–1567. [Google Scholar] [CrossRef] [Green Version]
- Somers, J.; Pöyry, T.; Willis, A.E. A perspective on mammalian upstream open reading frame function. Int. J. Biochem. Cell Biol. 2013, 45, 1690–1700. [Google Scholar] [CrossRef] [Green Version]
- Caliskan, N.; Peske, F.; Rodnina, M.V. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem. Sci. 2015, 40, 265–274. [Google Scholar] [CrossRef]
- Mikl, M.; Pilpel, Y.; Segal, E. High-throughput interrogation of programmed ribosomal frameshifting in human cells. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Belew, A.T.; Meskauskas, A.; Musalgaonkar, S.; Advani, V.M.; Sulima, S.O.; Kasprzak, W.K.; Shapiro, B.A.; Dinman, J.D. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nat. Cell Biol. 2014, 512, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.-Y.; Chang, K.-Y. An intermolecular RNA triplex provides insight into structural determinants for the pseudoknot stimulator of −1 ribosomal frameshifting. Nucleic Acids Res. 2009, 38, 1676–1685. [Google Scholar] [CrossRef]
- Chou, M.-Y.; Lin, S.-C.; Chang, K.-Y. Stimulation of -1 programmed ribosomal frameshifting by a metabolite-responsive RNA pseudoknot. RNA 2010, 16, 1236–1244. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.-H.; Luo, J.; Iwata-Reuyl, D.; Olsthoorn, R.C.L. Exploiting preQ1 Riboswitches to Regulate Ribosomal Frameshifting. ACS Chem. Biol. 2013, 8, 733–740. [Google Scholar] [CrossRef]
- Cho, C.-P.; Lin, S.-C.; Chou, M.-Y.; Hsu, H.-T.; Chang, K.-Y. Regulation of Programmed Ribosomal Frameshifting by Co-Translational Refolding RNA Hairpins. PLoS ONE 2013, 8, e62283. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.-T.; Lin, Y.-H.; Chang, K.-Y. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014, 42, 14070–14082. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Chang, K.Y. Rational design of a synthetic mammalian riboswitch as a ligand-responsive -1 ribosomal frame-shifting stimulator. Nucleic Acids Res. 2016, 44, 9005–9015. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, S.; Caliskan, N.; Rodnina, M.V.; Murata, A.; Nakatani, K. Small synthetic molecule-stabilized RNA pseudoknot as an activator for –1 ribosomal frameshifting. Nucleic Acids Res. 2018, 46, 8079–8089. [Google Scholar] [CrossRef]
- Nakatani, K.; Sando, S.; Saito, I. Scanning of guanine–guanine mismatches in DNA by synthetic ligands using surface plasmon resonance. Nat. Biotechnol. 2001, 19, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Setten, R.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Mueller, C. Design of AAV Vectors for Delivery of RNAi. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: New York, NY, USA, 2019; pp. 3–18. [Google Scholar]
- Atanasov, J.; Groher, F.; Weigand, J.E.; Suess, B. Design and implementation of a synthetic pre-miR switch for controlling miRNA biogenesis in mammals. Nucleic Acids Res. 2017, 45, e181. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-W.; Tseng, Y.-W.; Shen, C.-C.; Hsu, M.-N.; Hwu, J.-R.; Chang, C.-W.; Yeh, C.-J.; Chou, M.-Y.; Wu, J.-C.; Hu, Y.-C. Synthetic switch-based baculovirus for transgene expression control and selective killing of hepatocellular carcinoma cells. Nucleic Acids Res. 2018, 46, e93. [Google Scholar] [CrossRef]
- Matsuura, S.; Ono, H.; Kawasaki, S.; Kuang, Y.; Fujita, Y.; Saito, H. Synthetic RNA-based logic computation in mammalian cells. Nat. Commun. 2018, 9, 4847. [Google Scholar] [CrossRef] [PubMed]
- An, C.-I.; Trinh, V.B.; Yokobayashi, Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA 2006, 12, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Beisel, C.L.; Bayer, T.S.; Hoff, K.G.; Smolke, C.D. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 2008, 4, 224. [Google Scholar] [CrossRef]
- Beisel, C.L.; Chen, Y.Y.; Culler, S.J.; Hoff, K.G.; Smolke, C.D. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2010, 39, 2981–2994. [Google Scholar] [CrossRef]
- Beisel, C.L.; Bloom, R.J.; Smolke, C.D. Construction of Ligand-Responsive MicroRNAs that Operate Through Inhibition of Drosha Processing. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: New York, NY, USA, 2014; Volume 1111, pp. 259–267. [Google Scholar]
- Kumar, D.; An, C.-I.; Yokobayashi, Y. Conditional RNA Interference Mediated by Allosteric Ribozyme. J. Am. Chem. Soc. 2009, 131, 13906–13907. [Google Scholar] [CrossRef]
- Wong, R.S.; Chen, Y.Y.; Smolke, C.D. Regulation of T cell proliferation with drug-responsive microRNA switches. Nucleic Acids Res. 2018, 46, 1541–1552. [Google Scholar] [CrossRef] [Green Version]
- Pollak, N.M.; Cooper-White, J.J.; Macdonald, J. Translational control of enzyme scavenger expression with toxin-induced micro RNA switches. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Mou, H.; Zhong, G.; Gardner, M.R.; Wang, H.; Wang, Y.-W.; Cheng, D.; Farzan, M. Conditional Regulation of Gene Expression by Ligand-Induced Occlusion of a MicroRNA Target Sequence. Mol. Ther. 2018, 26, 1277–1286. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G.; Wang, H.; He, W.; Li, Y.; Mou, H.; Tickner, Z.J.; Tran, M.H.; Ou, T.; Yin, Y.; Diao, H.; et al. A reversible RNA on-switch that controls gene expression of AAV-delivered therapeutics in vivo. Nat. Biotechnol. 2020, 38, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Win, M.N.; Smolke, C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA 2007, 104, 14283–14288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittmann, A.; Suess, B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. BioSyst. 2011, 7, 2419–2427. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R. Control of gene expression by a natural metabolite-responsive ribozyme. Nat. Cell Biol. 2004, 428, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Ausländer, S.; Ketzer, P.; Hartig, J.S. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. BioSyst. 2010, 6, 807–814. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Y.; Jensen, M.C.; Smolke, C.D. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. USA 2010, 107, 8531–8536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobori, S.; Takahashi, K.; Yokobayashi, Y. Deep Sequencing Analysis of Aptazyme Variants Based on a Pistol Ribozyme. ACS Synth. Biol. 2017, 6, 1283–1288. [Google Scholar] [CrossRef] [Green Version]
- Mustafina, K.; Fukunaga, K.; Yokobayashi, Y. Design of Mammalian ON-Riboswitches Based on Tandemly Fused Aptamer and Ribozyme. ACS Synth. Biol. 2019, 9, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Piganeau, N.; Jenne, A.; Thuillier, V.; Famulok, M. An Allosteric Ribozyme Regulated by Doxycyline. Angew. Chem. Int. Ed. 2000, 39, 4369–4373. [Google Scholar] [CrossRef]
- Porter, E.B.; Polaski, J.T.; Morck, M.M.; Batey, R.T. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat. Chem. Biol. 2017, 13, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Beilstein, K.; Wittmann, A.; Grez, M.; Suess, B. Conditional Control of Mammalian Gene Expression by Tetracycline-Dependent Hammerhead Ribozymes. ACS Synth. Biol. 2015, 4, 526–534. [Google Scholar] [CrossRef]
- Kertsburg, A. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 2002, 30, 4599–4606. [Google Scholar] [CrossRef] [Green Version]
- Beaudoin, J.-D.; Perreault, J.-P. Potassium ions modulate a G-quadruplex-ribozyme’s activity. RNA 2008, 14, 1018–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouleau, S.G.; Jodoin, R.; Bisaillon, M.; Perreault, J.-P. Programming a Highly Structured Ribozyme into Complex Allostery Using RNA Oligonucleotides. ACS Chem. Biol. 2012, 7, 1802–1806. [Google Scholar] [CrossRef]
- Nomura, Y.; Zhou, L.; Miu, A.; Yokobayashi, Y. Controlling Mammalian Gene Expression by Allosteric Hepatitis Delta Virus Ribozymes. ACS Synth. Biol. 2013, 2, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Mamonkin, M.; Mukherjee, M.; Srinivasan, M.; Sharma, S.; Gomes-Silva, D.; Mo, F.; Krenciute, G.; Orange, J.S.; Brenner, M.K. Reversible Transgene Expression Reduces Fratricide and Permits 4-1BB Costimulation of CAR T Cells Directed to T-cell Malignancies. Cancer Immunol. Res. 2018, 6, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, W.; Huang, B.; Xu, S.; Li, Y.; Zhu, L.; Wu, Z.; Wu, X. AAV-Mediated In vivo CAR Gene Therapy for Targeting Human T Cell Leukemia. BioRxiv 2021, 4, ysy022. [Google Scholar] [CrossRef]
- Reid, C.; Nettesheim, E.R.; Connor, T.B.; Lipinski, D.M. Development of an inducible anti-VEGF rAAV gene therapy strategy for the treatment of wet AMD. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomura, Y.; Chien, H.-C.; Yokobayashi, Y. Direct screening for ribozyme activity in mammalian cells. Chem. Commun. 2017, 53, 12540–12543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felletti, M.; Stifel, J.; Wurmthaler, L.A.; Geiger, S.; Hartig, J.S. Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nat. Commun. 2016, 7, 12834. [Google Scholar] [CrossRef] [Green Version]
- Stieger, K.; Belbellaa, B.; Le Guiner, C.; Moullier, P.; Rolling, F. In vivo gene regulation using tetracycline-regulatable systems. Adv. Drug Deliv. Rev. 2009, 61, 527–541. [Google Scholar] [CrossRef]
- Wurmthaler, L.A.; Klauser, B.; Hartig, J.S. Highly motif- and organism-dependent effects of naturally occurring hammerhead ribozyme sequences on gene expression. RNA Biol. 2017, 15, 231–241. [Google Scholar] [CrossRef]
- Fritz, J.J.; White, D.A.; Lewin, A.S.; Hauswirth, W.W. Designing and characterizing hammerhead ribozymes for use in AAV vector-mediated retinal gene therapies. Methods Enzymol. 2002, 346, 358–377. [Google Scholar] [CrossRef]
- Yen, L.; Svendsen, J.; Lee, J.-S.; Gray, J.T.; Magnier, M.; Baba, T.; D’Amato, R.J.; Mulligan, R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nat. Cell Biol. 2004, 431, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Morcos, P.A.; Li, Y.; Jiang, S. Vivo-Morpholinos: A non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. BioTechniques 2008, 45, 613–623. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef] [PubMed]
- Wadman, M. Promising drug for Huntington disease fails in major trial. Science 2021. [Google Scholar] [CrossRef]
- Zhong, G.; Wang, H.; Bailey, C.C.; Gao, G.; Farzan, M. Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. eLife 2016, 5, e18858. [Google Scholar] [CrossRef] [PubMed]
- Strobel, B.; Düchs, M.J.; Blazevic, D.; Rechtsteiner, P.; Braun, C.; Baum-Kroker, K.S.; Schmid, B.; Ciossek, T.; Gottschling, D.; Hartig, J.S.; et al. A Small-Molecule-Responsive Riboswitch Enables Conditional Induction of Viral Vector-Mediated Gene Expression in Mice. ACS Synth. Biol. 2020, 9, 1292–1305. [Google Scholar] [CrossRef]
- Ausländer, S.; Stücheli, P.; Rehm, C.; Ausländer, D.; Hartig, J.S.; Fussenegger, M. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Chem. Biol. 2014, 11, 1154–1160. [Google Scholar] [CrossRef]
- Robertson, M.P.; Ellington, A. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999, 17, 62–66. [Google Scholar] [CrossRef]
- Koizumi, M.; Soukup, G.A.; Kerr, J.N.; Breaker, R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Genet. 1999, 6, 1062–1071. [Google Scholar] [CrossRef]
- Ferguson, A.; Boomer, R.M.; Kurz, M.; Keene, S.C.; Diener, J.L.; Keefe, A.D.; Wilson, C.; Cload, S.T. A novel strategy for selection of allosteric ribozymes yields RiboReporter™ sensors for caffeine and aspartame. Nucleic Acids Res. 2004, 32, 1756–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piganeau, N.; Thuillier, V.; Famulok, M. In vitro selection of allosteric ribozymes: Theory and experimental validation. J. Mol. Biol. 2001, 312, 1177–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, H.; Furukawa, K.; Breaker, R.R. Engineered Allosteric Ribozymes That Sense the Bacterial Second Messenger Cyclic Diguanosyl 5′-Monophosphate. Anal. Chem. 2012, 84, 4935–4941. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, K.; Gu, H.; Breaker, R.R. In Vitro Selection of Allosteric Ribozymes that Sense the Bacterial Second Messenger c-di-GMP. In Artificial Riboswitches: Methods and Protocols; Humana Press: New York, NY, USA, 2014; Volume 1111, pp. 209–220. [Google Scholar]
- Kobori, S.; Nomura, Y.; Miu, A.; Yokobayashi, Y. High-throughput assay and engineering of self-cleaving ribozymes by sequencing. Nucleic Acids Res. 2015, 43, e85. [Google Scholar] [CrossRef]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goler, J.A.; Carothers, J.M.; Keasling, J.D. Dual-Selection for Evolution of In Vivo Functional Aptazymes as Riboswitch Parts. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: New York, NY, USA, 2014; Volume 1111, pp. 221–235. [Google Scholar]
- Wieland, M.; Hartig, J.S. Improved Aptazyme Design and In Vivo Screening Enable Riboswitching in Bacteria. Angew. Chem. Int. Ed. 2008, 47, 2604–2607. [Google Scholar] [CrossRef]
- Muranaka, N.; Sharma, V.; Nomura, Y.; Yokobayashi, Y. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res. 2009, 37, e39. [Google Scholar] [CrossRef] [Green Version]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Stifel, J.; Spöring, M.; Hartig, J.S. Expanding the toolbox of synthetic riboswitches with guanine-dependent aptazymes. Synth. Biol. 2019, 4, ysy022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, J.; Cheng, H.; Sun, Y.; Liu, M.; Wu, Z.; Pei, R. Conditional control of suicide gene expression in tumor cells with theophylline-responsive ribozyme. Gene Ther. 2016, 24, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Zhou, S.; Huang, X.; Yuan, Y.; Du, F.; Dong, J.; Chen, G.; Cui, X.; Tang, Z. Intracellular Selection of Theophylline-Sensitive Hammerhead Aptazyme. Mol. Ther. Nucleic Acids 2020, 20, 400–408. [Google Scholar] [CrossRef]
- Xiang, J.S.; Kaplan, M.; Dykstra, P.; Hinks, M.; McKeague, M.; Smolke, C.D. Massively parallel RNA device engineering in mammalian cells with RNA-Seq. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Strobel, B.; Spöring, M.; Klein, H.; Blazevic, D.; Rust, W.; Sayols, S.; Hartig, J.S.; Kreuz, S. High-throughput identification of synthetic riboswitches by barcode-free amplicon-sequencing in human cells. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Townshend, B.; Xiang, J.S.; Manzanarez, G.; Hayden, E.J.; Smolke, C.D. A multiplexed, automated evolution pipeline enables scalable discovery and characterization of biosensors. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Zhan, H.; Ding, M.; Zhou, Q.; Li, A.; Cai, Z.; Huang, W.; Liu, Y. Improving transgene expression and CRISPR-Cas9 efficiency with molecular engineering-based molecules. Clin. Transl. Med. 2020, 10, e194. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibraheim, R.; Song, C.-Q.; Mir, A.; Amrani, N.; Xue, W.; Sontheimer, E.J. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 2018, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Swiech, L.; Heidenreich, M.; Banerjee, A.; Habib, N.; Li, Y.; Trombetta, J.J.; Sur, M.; Zhang, F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 102–106. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nat. Cell Biol. 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Collantes, J.C.; Tan, V.M.; Xu, H.; Ruiz-Urigüen, M.; Alasadi, A.; Guo, J.; Tao, H.; Su, C.; Tyc, K.M.; Selmi, T.; et al. Development and Characterization of a Modular CRISPR and RNA Aptamer Mediated Base Editing System. CRISPR J. 2021, 4, 58–68. [Google Scholar] [CrossRef]
- Carlson-Stevermer, J.; Abdeen, A.A.; Kohlenberg, L.; Goedland, M.; Molugu, K.; Lou, M.; Saha, K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Su, J.-H.; Zhang, F.; Zhuang, X. An RNA-aptamer-based two-color CRISPR labeling system. Sci. Rep. 2016, 6, 26857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Stanford, W.; De Solis, C.; Aradhana; Abraham, N.D.; Dao, T.-M.J.; Thaseen, S.; Sairavi, A.; Gonzalez, C.U.; Ploski, J.E. The Development of an AAV-Based CRISPR SaCas9 Genome Editing System That Can Be Delivered to Neurons in vivo and Regulated via Doxycycline and Cre-Recombinase. Front. Mol. Neurosci. 2018, 11, 413. [Google Scholar] [CrossRef] [Green Version]
- Kundert, K.; Lucas, J.E.; Watters, K.E.; Fellmann, C.; Ng, A.H.; Heineike, B.; Fitzsimmons, C.M.; Oakes, B.L.; Qu, J.; Prasad, N.; et al. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, R.S.; Ozdilek, B.A.; Garst, A.D.; Choudhury, A.; Batey, R.T. Small molecule regulated sgRNAs enable control of genome editing in E. coli by Cas9. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lin, B.; An, Y.; Meng, L.; Zhang, H.; Song, J.; Zhu, Z.; Liu, W.; Song, Y.; Yang, C. Control of CRISPR-Cas9 with small molecule-activated allosteric aptamer regulating sgRNAs. Chem. Commun. 2019, 55, 12223–12226. [Google Scholar] [CrossRef]
- Liu, Y.; Zhan, Y.; Chen, Z.; He, A.; Li, J.; Wu, H.; Liu, L.; Zhuang, C.; Lin, J.; Guo, X.; et al. Directing cellular information flow via CRISPR signal conductors. Nat. Methods 2016, 13, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Hu, J.H.; Liu, D.R. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017, 8, 15939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Liu, Y.; Lai, P.; Ye, H.; Xu, L. Conditional guide RNA through two intermediate hairpins for programmable CRISPR/Cas9 function: Building regulatory connections between endogenous RNA expressions. Nucleic Acids Res. 2020, 48, 11773–11784. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Zhuang, C.; Zhou, Q.; Huang, X.; Gui, Y.; Lai, Y.; Yang, S. Engineered CRISPR/Cas13d Sensing hTERT Selectively Inhibits the Progression of Bladder Cancer In Vitro. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef]
- Nathwani, A.C.; Rosales, C.; McIntosh, J.; Rastegarlari, G.; Nathwani, D.; Raj, D.; Nawathe, S.; Waddington, S.N.; Bronson, R.; Jackson, S.; et al. Long-term Safety and Efficacy Following Systemic Administration of a Self-complementary AAV Vector Encoding Human FIX Pseudotyped With Serotype 5 and 8 Capsid Proteins. Mol. Ther. 2011, 19, 876–885. [Google Scholar] [CrossRef]
- Li, A.; Lee, C.; Hurley, A.E.; Jarrett, K.E.; De Giorgi, M.; Lu, W.; Balderrama, K.S.; Doerfler, A.M.; Deshmukh, H.; Ray, A.; et al. A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol. Ther. Methods Clin. Dev. 2019, 12, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Farzan, M. A Safety Switch for an Effective HIV-1 Vaccine. Available online: https://grantome.com/grant/NIH/DP1-DA043912-01 (accessed on 7 May 2021).
- Gu, H.; Furukawa, K.; Weinberg, Z.; Berenson, D.F.; Breaker, R.R. Small, Highly Active DNAs That Hydrolyze DNA. J. Am. Chem. Soc. 2013, 135, 9121–9129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, M.; Sachdeva, A.; Silverman, S.K. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 2009, 5, 718–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velez, T.E.; Singh, J.; Xiao, Y.; Allen, E.C.; Wong, O.Y.; Chandra, M.; Kwon, S.C.; Silverman, S.K. Systematic Evaluation of the Dependence of Deoxyribozyme Catalysis on Random Region Length. ACS Comb. Sci. 2012, 14, 680–687. [Google Scholar] [CrossRef] [Green Version]
- Carmi, N.; Balkhi, S.R.; Breaker, R. Cleaving DNA with DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 2233–2237. [Google Scholar] [CrossRef] [Green Version]
- Gysbers, R.; Tram, K.; Gu, J.; Li, Y. Evolution of an Enzyme from a Noncatalytic Nucleic Acid Sequence. Sci. Rep. 2015, 5, 11405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yang, J.; Yuan, X.; Cao, J.; Xu, J.; Chaput, J.C.; Li, Z.; Yu, H. A Novel Small RNA-Cleaving Deoxyribozyme with a Short Binding Arm. Sci. Rep. 2019, 9, 8224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-B.; Kong, R.-M.; Lu, Y. Metal Ion Sensors Based on DNAzymes and Related DNA Molecules. Annu. Rev. Anal. Chem. 2011, 4, 105–128. [Google Scholar] [CrossRef] [Green Version]
- Isaka, Y. DNAzymes as potential therapeutic molecules. Curr. Opin. Mol. Ther. 2007, 9, 132–136. [Google Scholar]
- Egrimpe, B. Deoxyribozymes: New Therapeutics to Treat Central Nervous System Disorders. Front. Mol. Neurosci. 2011, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Adornetto, G.; Porchetta, A.; Palleschi, G.; Plaxco, K.W.; Ricci, F. A general approach to the design of allosteric, transcription factor-regulated DNAzymes. Chem. Sci. 2015, 6, 3692–3696. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Simon, A.J.; Pei, H.; Shi, J.; Li, J.; Huang, Q.; Plaxco, K.W.; Fan, C. Activity modulation and allosteric control of a scaffolded DNAzyme using a dynamic DNA nanostructure. Chem. Sci. 2015, 7, 1200–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossetti, M.; Ranallo, S.; Idili, A.; Palleschi, G.; Porchetta, A.; Ricci, F. Allosteric DNA nanoswitches for controlled release of a molecular cargo triggered by biological inputs. Chem. Sci. 2016, 8, 914–920. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Lee, C.H.; Han, S.R.; Lee, S.-W. Therapeutic Applications of Aptamer-Based Riboswitches. Nucleic Acid Ther. 2016, 26, 44–51. [Google Scholar] [CrossRef]
- Bunn, H.F. Erythropoietin. Cold Spring Harb. Perspect. Med. 2013, 3, a011619. [Google Scholar] [CrossRef] [Green Version]
- Rivera, V.M.; Gao, G.-P.; Grant, R.L.; Schnell, M.A.; Zoltick, P.W.; Rozamus, L.W.; Clackson, T.; Wilson, J. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood 2005, 105, 1424–1430. [Google Scholar] [CrossRef]
- Tao, Y.; Zhu, Q.; Wang, L.; Zha, X.; Teng, D.; Xu, L. Adeno-associated virus (AAV)-mediated neuroprotective effects on the degenerative retina: The therapeutic potential of erythropoietin. Fundam. Clin. Pharmacol. 2020, 34, 131–147. [Google Scholar] [CrossRef]
- Xue, Y.-Q.; Ma, B.-F.; Zhao, L.-R.; Tatom, J.B.; Li, B.; Jiang, L.-X.; Klein, R.L.; Duan, W.-M. AAV9-mediated erythropoietin gene delivery into the brain protects nigral dopaminergic neurons in a rat model of Parkinson’s disease. Gene Ther. 2009, 17, 83–94. [Google Scholar] [CrossRef]
- Johnston, J.; Tazelaar, J.; Rivera, V.M.; Clackson, T.; Gao, G.-P.; Wilson, J.M. Regulated expression of erythropoietin from an AAV vector safely improves the anemia of β-thalassemia in a mouse model. Mol. Ther. 2003, 7, 493–497. [Google Scholar] [CrossRef]
- Samakoglu, S.; Bohl, D.; Heard, J.M. Mechanisms Leading to Sustained Reversion of β-Thalassemia in Mice by Doxycycline-Controlled Epo Delivery from Muscles. Mol. Ther. 2002, 6, 793–803. [Google Scholar] [CrossRef]
- Gao, G.; Lebherz, C.; Weiner, D.J.; Grant, R.; Calcedo, R.; McCullough, B.; Bagg, A.; Zhang, Y.; Wilson, J.M. Erythropoietin gene therapy leads to autoimmune anemia in macaques. Blood 2004, 103, 3300–3302. [Google Scholar] [CrossRef] [Green Version]
- Sebestyen, M. Protein-free Regulation of Erythropoietin Expression by a Drug-Sensing Riboswitch. Available online: https://www.sbir.gov/sbirsearch/detail/236751 (accessed on 7 May 2021).
- Boyne, A.R.; Danos, O.F.; Volles, M.J.; Guo, X. Regulation of Gene Expression by Aptamer-Mediated Modulation of Alternative Splicing. Available online: https://patents.google.com/patent/US20200087683A1/en (accessed on 7 May 2021).
- Sanders, J.W.; Ponzio, T.A. Vectored immunoprophylaxis: An emerging adjunct to traditional vaccination. Trop. Dis. Travel Med. Vaccines 2017, 3, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Burton, D.R. Advancing an HIV vaccine; advancing vaccinology. Nat. Rev. Immunol. 2019, 19, 77–78. [Google Scholar] [CrossRef]
- Gulick, R.M.; Flexner, C. Long-Acting HIV Drugs for Treatment and Prevention. Annu. Rev. Med. 2019, 70, 137–150. [Google Scholar] [CrossRef]
- Niessl, J.; Baxter, A.E.; Mendoza, P.; Jankovic, M.; Cohen, Y.Z.; Butler, A.L.; Lu, C.-L.; Dube, M.; Shimeliovich, I.; Grüll, H.; et al. Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nat. Med. 2020, 26, 222–227. [Google Scholar] [CrossRef]
- Lewis, A.D.; Chen, R.; Montefiori, D.C.; Johnson, P.R.; Clark, K.R. Generation of Neutralizing Activity against Human Immunodeficiency Virus Type 1 in Serum by Antibody Gene Transfer. J. Virol. 2002, 76, 8769–8775. [Google Scholar] [CrossRef] [Green Version]
- Balazs, A.B.; Bloom, J.; Hong, C.M.; Rao, D.; Baltimore, D. Broad protection against influenza infection by vectored immunoprophylaxis in mice. Nat. Biotechnol. 2013, 31, 647–652. [Google Scholar] [CrossRef] [PubMed]
- De Jong, Y.P.; Dorner, M.; Mommersteeg, M.; Xiao, J.W.; Balazs, A.B.; Robbins, J.B.; Winer, B.Y.; Gerges, S.; Vega, K.; Labitt, R.N.; et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci. Transl. Med. 2014, 6, 254ra129. [Google Scholar] [CrossRef] [Green Version]
- Skaricic, D.; Traube, C.; De, B.; Joh, J.; Boyer, J.; Crystal, R.G.; Worgall, S. Genetic delivery of an anti-RSV antibody to protect against pulmonary infection with RSV. Virology 2008, 378, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flingai, S.; Plummer, E.M.; Patel, A.; Shresta, S.; Mendoza, J.M.; Broderick, K.E.; Sardesai, N.Y.; Muthumani, K.; Weiner, D.B. Protection against dengue disease by synthetic nucleic acid antibody prophylaxis/immunotherapy. Sci. Rep. 2015, 5, 12616. [Google Scholar] [CrossRef]
- Muthumani, K.; Block, P.; Flingai, S.; Muruganantham, N.; Chaaithanya, I.K.; Tingey, C.; Wise, M.; Reuschel, E.L.; Chung, C.; Muthumani, A.; et al. Rapid and Long-Term Immunity Elicited by DNA-Encoded Antibody Prophylaxis and DNA Vaccination Against Chikungunya Virus. J. Infect. Dis. 2016, 214, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rghei, A.D.; Van Lieshout, L.P.; Santry, L.A.; Guilleman, M.M.; Thomas, S.P.; Susta, L.; Karimi, K.; Bridle, B.W.; Wootton, S.K. AAV Vectored Immunoprophylaxis for Filovirus Infections. Trop. Med. Infect. Dis. 2020, 5, 169. [Google Scholar] [CrossRef]
- De, B.P.; Hackett, N.R.; Crystal, R.G.; Boyer, J.L. Rapid/Sustained Anti-anthrax Passive Immunity Mediated by Co-administration of Ad/AAV. Mol. Ther. 2008, 16, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Deal, C.; Balazs, A.B.; Espinosa, D.A.; Zavala, F.; Baltimore, D.; Ketner, G. Vectored antibody gene delivery protects against Plasmodium falciparum sporozoite challenge in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 12528–12532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priddy, F.H.; Lewis, D.J.M.; Gelderblom, H.C.; Hassanin, H.; Streatfield, C.; LaBranche, C.; Hare, J.; Cox, J.; Dally, L.; Bendel, D.; et al. Adeno-associated virus vectored immunoprophylaxis to prevent HIV in healthy adults: A phase 1 randomised controlled trial. Lancet HIV 2019, 6, e230–e239. [Google Scholar] [CrossRef] [Green Version]
- Mingozzi, F.; Hasbrouck, N.C.; Basner-Tschakarjan, E.; Edmonson, S.A.; Hui, D.J.; Sabatino, D.E.; Zhou, S.; Wright, J.F.; Jiang, H.; Pierce, G.F.; et al. Modulation of tolerance to the transgene product in a nonhuman primate model of AAV-mediated gene transfer to liver. Blood 2007, 110, 2334–2341. [Google Scholar] [CrossRef] [Green Version]
- Martino, A.T.; Suzuki, M.; Markusic, D.M.; Zolotukhin, I.; Ryals, R.C.; Moghimi, B.; Ertl, H.C.J.; Muruve, D.A.; Lee, B.; Herzog, R.W. The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9–dependent innate immune responses in the liver. Blood 2011, 117, 6459–6468. [Google Scholar] [CrossRef] [Green Version]
- Markusic, D.; Herzog, R.W. Hepatic Gene Transfer of Factor IX Reverses Inhibitors and Protects From Anaphylaxis in a Murine Hemophilia B Model. Blood 2011, 118, 669. [Google Scholar] [CrossRef]
- Gardner, M.R.; Kattenhorn, L.M.; Kondur, H.R.; von Schaewen, M.; Dorfman, T.; Chiang, J.J.; Haworth, K.G.; Decker, J.M.; Alpert, M.D.; Bailey, C.C.; et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nat. Cell Biol. 2015, 519, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Mingozzi, F.; High, K.A. Immune responses to AAV vectors: Overcoming barriers to successful gene therapy. Blood 2013, 122, 23–36. [Google Scholar] [CrossRef]
- Herzog, R.W.; Mount, J.D.; Arruda, V.R.; High, K.A.; Lothrop, C.D., Jr. Muscle-Directed Gene Transfer and Transient Immune Suppression Result in Sustained Partial Correction of Canine Hemophilia B Caused by a Null Mutation. Mol. Ther. 2001, 4, 192–200. [Google Scholar] [CrossRef]
- Gardner, M.R. Promise and Progress of an HIV-1 Cure by Adeno-Associated Virus Vector Delivery of Anti-HIV-1 Biologics. Front. Cell. Infect. Microbiol. 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Defense Advanced Research Projects Agency. Dialing Up the Body’s Defenses Against Public Health and National Security Threats. Available online: https://www.darpa.mil/news-events/2018-05-25 (accessed on 7 May 2021).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tickner, Z.J.; Farzan, M. Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals 2021, 14, 554. https://doi.org/10.3390/ph14060554
Tickner ZJ, Farzan M. Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals. 2021; 14(6):554. https://doi.org/10.3390/ph14060554
Chicago/Turabian StyleTickner, Zachary J., and Michael Farzan. 2021. "Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors" Pharmaceuticals 14, no. 6: 554. https://doi.org/10.3390/ph14060554
APA StyleTickner, Z. J., & Farzan, M. (2021). Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals, 14(6), 554. https://doi.org/10.3390/ph14060554