Biotransformation of Modified Benzylisoquinoline Alkaloids: Boldine and Berberine and In Silico Molecular Docking Studies of Metabolites on Telomerase and Human Protein Tyrosine Phosphatase 1B
Abstract
1. Introduction
2. Results and Discussion
2.1. The Biotransformation Products Identification
2.2. Molecular Docking Investigations
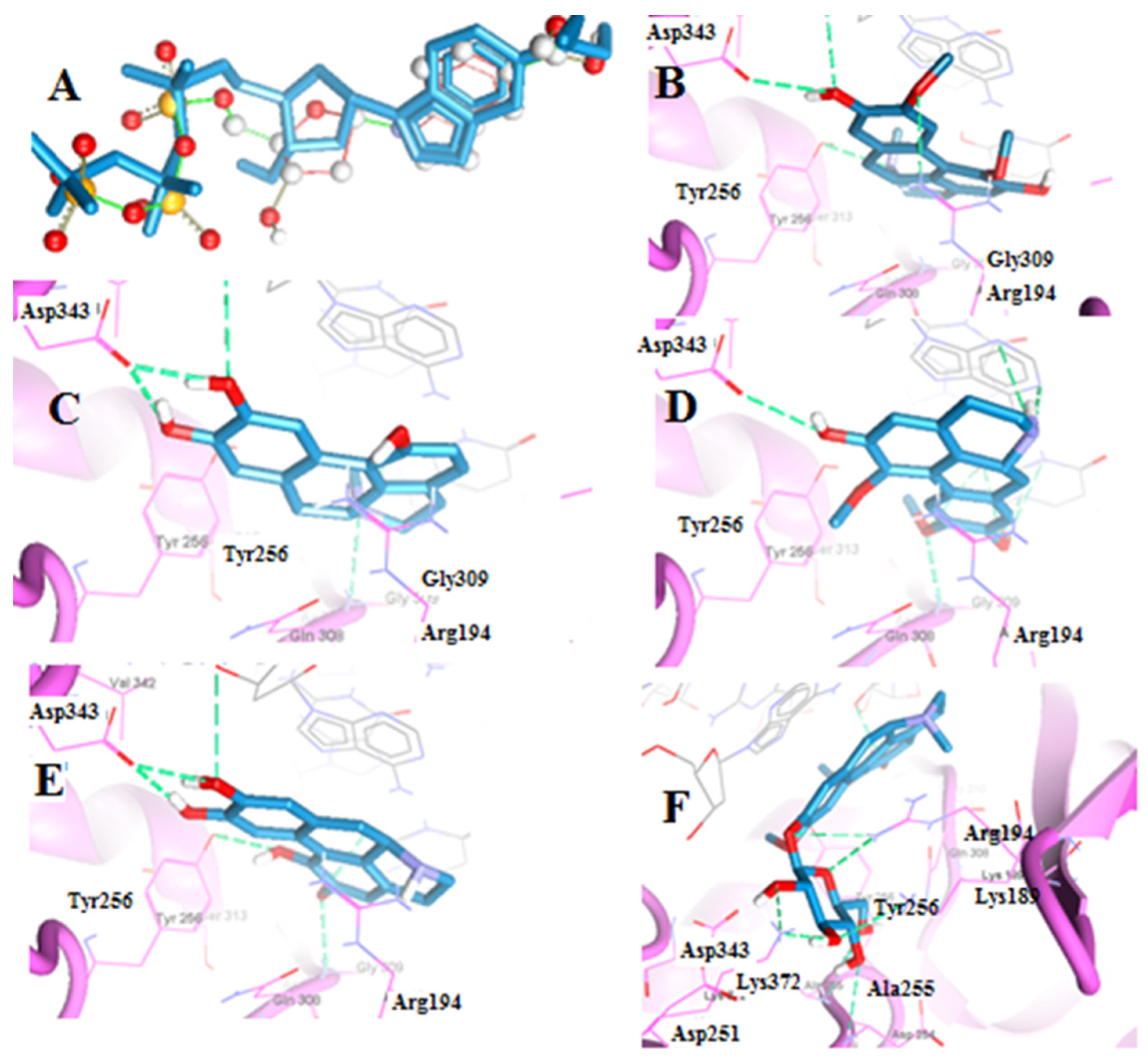
2.2.1. Molecular Docking of Boldine Metabolites to Telomerase Enzyme
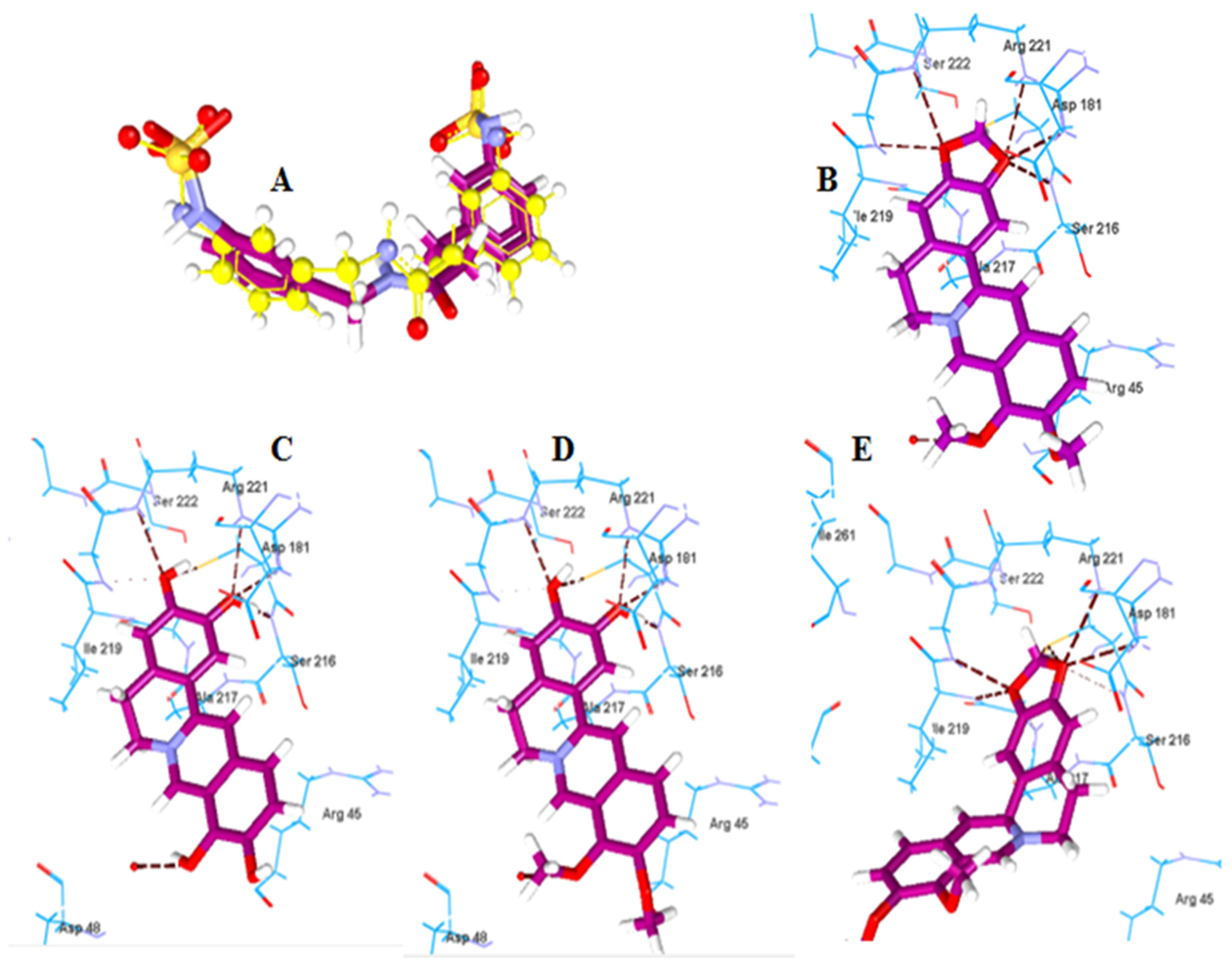
2.2.2. Molecular Docking of Berberine Metabolites to the Protein Tyrosine Phosphatase 1B (PTP-1B)
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Chemicals
3.3. Microorganisms
3.4. Preparative Scale Fermentation and Purification of the Metabolites
3.4.1. Boldine Transformation by Cunninghamella blackesleeana NRRL 1369
3.4.2. Boldine Transformation by Cunninghamella blackesleeana MR 198
3.4.3. Boldine Transformation by Penicillium chrysogeneum ATCC 10002
3.4.4. Berberine Transformation by Cunninghamella blackesleeana NRRL 1369
3.4.5. Berberine Transformation by Penicillium chrysogeneum ATCC 10002
3.4.6. Berberine Transformation by Cunninghamella elegans NRRL 2310
3.5. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smitha, M.S.; Singh, S.; Singh, R. Microbial bio transformation: A process for chemical alterations. J. Bacteriol. Mycol. 2017, 4, 47–51. [Google Scholar]
- Rathbone, D.A.; Bruce, N.C. Microbial transformation of alkaloids. Curr. Opin. Microbiol. 2002, 5, 274–281. [Google Scholar] [CrossRef]
- Speisky, H.; Cassels, B.K. Boldo and boldine: An emerging case of natural drug development. Pharmacol. Res. 1994, 29, 1–12. [Google Scholar] [CrossRef]
- Chen, K.; Cheng, P.; Zhu, W.T.; Liang, S.; Yang, Q.; Parkman, V.A.; Zhou, C.H.; Jing, X.Z.; Liu, H.; Wang, Y.T.; et al. Boldine Ameliorates Estrogen Deficiency-Induced Bone Loss via Inhibiting Bone Resorption. Front. Pharmacol. 2018, 13, 1046. [Google Scholar] [CrossRef]
- Lau, Y.S.; Ling, W.C.; Murugan, D.; Mustafa, M.R. Boldine Ameliorates Vascular Oxidative Stress and Endothelial Dysfunction: Therapeutic Implication for Hypertension and Diabetes. J. Cardiovasc. Pharmacol. 2015, 65, 522–531. [Google Scholar] [CrossRef]
- Boeing, T.; Mariano, L.; Dos Santos, A.C.; Tolentino, B.; Vargas, A.C.; de Souza, P.; Nesello, L.; da Silva, L.M. Gastroprotective effect of the alkaloid boldine: Involvement of non-protein sulfhydryl groups, prostanoids and reduction on oxidative stress. Chem.-Biol. Interact. 2020, 25, 109166. [Google Scholar] [CrossRef]
- Shuker, E.; Farhood, M.; Al-Qudaihi, G.; Fouad, D. Potential Effects of Boldine on Oxidative Stress, Apoptosis, and Inflammatory Changes Induced by the Methylprednisolone Hepatotoxicity in Male Wistar Rats. Dose Response Int. J. 2022, 20, 155932582210828. [Google Scholar] [CrossRef]
- Backhouse, N.; Delporte, C.; Givernau, M.; Cassels, B.K.; Valenzuela, A.; Speisky, H. Anti-inflammatory and antipyretic effects of boldine. Agents Actions 1994, 42, 114–117. [Google Scholar] [CrossRef]
- Kara, E.; Kahraman, E.; Dayar, E.; Anacak, G.; Demir, O.; Gidener, S.; Atabey, N.; Durmus, N. The role of resistin on metabolic syndrome-induced erectile dysfunction and the possible therapeutic effect of Boldine. Andrology 2020, 8, 1728–1735. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The Anti-Cancer Mechanisms of Berberine: A Review. Cancer Manag. Res. 2020, 30, 695–702. [Google Scholar] [CrossRef]
- Xu, X.; Yi, H.; Wu, J.; Kuang, T.; Zhang, J.; Li, Q.; Du, H.; Xu, T.; Jiang, G.; Fan, G. Therapeutic effect of berberine on metabolic diseases: Both pharmacological data and clinical evidence. Biomed. Pharmacother. 2021, 1, 110984. [Google Scholar] [CrossRef]
- Leng, S.H.; Lu, F.E.; Xu, L.J. Therapeutic effects of berberine in impaired glucose tolerance rats and its influence on insulin secretion. Acta Pharmacol. Sin. 2004, 25, 496–502. [Google Scholar]
- Lu, Y.; Huang, J.; Zhang, Y.; Huang, Z.; Yan, W.; Zhou, T.; Wang, Z.; Liao, L.; Cao, H.; Tan, B. Therapeutic Effects of Berberine Hydrochloride on Stress-Induced Diarrhea-Predominant Irritable Bowel Syndrome Rats by Inhibiting Neurotransmission in Colonic Smooth Muscle. Front. Pharmacol. 2021, 14, 596686. [Google Scholar] [CrossRef]
- Bustanji, Y.; Taha, M.O.; Yousef, A.M.; Al-Bakri, G.A. Berberine potently inhibits protein tyrosine phosphatase 1B: Investigation by docking simulation and experimental validation. J. Enzym. Inhib. Med. Chem. 2006, 21, 163–171. [Google Scholar] [CrossRef]
- Saxena, A.K.; Pandey, G.; Gupta, S.; Singh, A.B.; Srivastava, A.K. Synthesis of protein tyrosine phosphatase 1B inhibitors: Model validation and docking studies. Bioorganic Med. Chem. Lett. 2009, 19, 2320–2323. [Google Scholar] [CrossRef]
- Davis, P.J.; Talaat, R.E. Microbiological Systems in Organic Synthesis: Preparative-Scale Resolution of (RS)-Glaucine by Fusarium solani and Stereospecific Oxidation of (R)-(−)-Glaucine by Aspergillus flavipes. Appl. Environ. Microbiol. 1981, 41, 1243–1247. [Google Scholar] [CrossRef]
- El-Aasr, M.; Eliwa, D.; Albadry, M.; Ibrahim, A.S.; Kabbash, A.; Meepagala, K.M.; Khan, I.A.; Khan, S.I.; Ross, S.A. Microbial transformation of some simple isoquinoline and benzylisoquinoline alkaloids and in vitro studies of their metabolites. Phytochemistry 2021, 189, 112828. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, J.; Tan, Y.; Feng, W.; Peng, C. Interactions between gut microbiota and berberine, a necessary procedure to understand the mechanisms of berberine. J. Pharm. Anal. 2021, 12, 541–555. [Google Scholar] [CrossRef]
- Wang, J.; Feng, W.; Tang, F.; Ao, H.; Peng, C. Gut microbial transformation, a potential improving factor in the therapeutic activities of four groups of natural compounds isolated from herbal medicines. Fitoterapia 2019, 138, 104293. [Google Scholar] [CrossRef]
- Feng, R.; Shou, J.W.; Zhao, Z.X.; He, C.Y.; Ma, C.; Huang, M.; Fu, J.; Tan, X.S.; Li, X.Y.; Wen, B.Y.; et al. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci. Rep. 2015, 15, 12155. [Google Scholar] [CrossRef]
- Li, Y.; Ren, G.; Wang, Y.X.; Kong, W.J.; Yang, P.; Wang, Y.M.; Li, Y.H.; Yi, H.; Li, Z.R.; Song, D.Q.; et al. Bioactivities of berberine metabolites after transformation through CYP450 isoenzymes. J. Transl. Med. 2011, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Yimlaz, D.; Gizem, F.; Akbulut, B. Curvularia lunata: A fungus for possible berberine transformation. Int. J. Second. Metab. 2022, 9, 66–73. [Google Scholar]
- Agusta, A.; Wulansari, D.; Tiwi, P.; Fathoni, A. Biotransformation of Protoberberine Alkaloids by the Endophytic Fungus Coelomycetes AFKR-3 Isolated from Yellow Moonsheed Plant (Archangelisia Flava (L.) Merr.). Procedia Chem. 2014, 13, 38–43. [Google Scholar] [CrossRef]
- Jiménez, I.; Speisky, H. Biological disposition of boldine: In Vitro and in vivo studies. Phytother. Res. 2000, 14, 254–260. [Google Scholar] [CrossRef]
- Buchanan, M.; Carroll, A.; Pass, D.; Quinn, R. Aporphine Alkaloids from the Chinese Tree Neolitsea Aurata Var. Paraciculata. Nat. Prod. Commun. 2007, 2, 1934578X0700200306. [Google Scholar] [CrossRef]
- Yu, L.L.; Li, R.T.; Ai, Y.B.; Liu, W.; Deng, Z.S.; Zou, Z.M. Protoberberine isoquinoline alkaloids from Arcangelisia gusanlung. Molecules 2014, 29, 13332–13341. [Google Scholar] [CrossRef]
- Wang, K.; Feng, X.; Chai, L.; Cao, S.; Qiu, F. The metabolism of berberine and its contribution to the pharmacological effects. Drug Metab. Rev. 2017, 49, 139–157. [Google Scholar] [CrossRef]
- Hroch, M.; Mičuda, S.; Cermanová, J.; Chládek, J.; Tomšík, P. Development of an HPLC fluorescence method for determination of boldine in plasma, bile and urine of rats and identification of its major metabolites by LC-MS/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 1, 48–56. [Google Scholar] [CrossRef]
- Zhang, Z.; Cong, L.; Peng, R.; Han, P.; Ma, S.R.; Pan, L.B.; Fu, J.; Yu, H.; Wang, Y.; Jiang, J.D. Transformation of berberine to its demethylated metabolites by the CYP51 enzyme in the gut microbiota. J. Pharm. Anal. 2021, 11, 628–637. [Google Scholar] [CrossRef]
- Moon, J.M.; Ratliff, K.M.; Hagele, A.M.; Stecker, R.A.; Mumford, P.W.; Kerksick, C.M. Absorption Kinetics of Berberine and Dihydroberberine and Their Impact on Glycemia: A Randomized, Controlled, Crossover Pilot Trial. Nutrients 2021, 14, 124. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, W.; Huang, W.; Plucinsky, B.; Garcia-Vazquez, N.; Robinson, N.J.; Schiemann, W.P.; Berdis, A.J.; Skordalakes, E.; Taylor, D.J. A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity. PLoS Biol. 2019, 17, e3000204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mar, V.; Zhou, W.; Harrington, L.; Robinson, M.O. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999, 13, 2388–2399. [Google Scholar] [CrossRef]
- Noureini, S.; Kheirabadi, M.; Masoumi, F.; Khosrogerdi, F.; Zarei, Y.; Suárez-Rozas, C.; Salas-Norambuena, J.; Cassels, B. Telomerase Inhibition by a New Synthetic Derivative of the Aporphine Alkaloid Boldine. Int. J. Mol. Sci. 2018, 19, 1239. [Google Scholar] [CrossRef] [PubMed]
- Eliwa, D.; Albadry, M.A.; Ibrahim, A.S.; Kabbash, A.; Meepagala, K.; Khan, I.A.; El-Aasr, M.; Ross, S.A. Biotransformation of papaverine and in silico docking studies of the metabolites on human phosphodiesterase 10a. Phytochemistry 2021, 183, 112598. [Google Scholar] [CrossRef]
- Klopfenstein, S.R.; Evdokimov, A.G.; Colson, A.O.; Fairweather, N.T.; Neuman, J.J.; Maier, M.B.; Gray, J.L.; Gerwe, G.S.; Stake, G.E.; Howard, B.W.; et al. 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors. Bioorganic Med. Chem. Lett. 2006, 16, 1574–1578. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.S.; Ragab, A.E. New adipate esters from Cunninghamella echinulata: Isolation, identification, biosynthesis and in silico prediction of potential opioid/anti-opioid and antidiabetic activities. Nat. Prod. Res. 2022, 19, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ragab, A.E.; Badawy, E.T.; Aboukhatwa, S.M.; Abdel-Aziz, M.M.; Kabbash, A.; Abo Elseoud, K.A. Isonicotinic acid N-oxide, from isoniazid biotransformation by Aspergillus niger, as an InhA inhibitor antituberculous agent against multiple and extensively resistant strains supported by in silico docking and ADME prediction. Nat. Prod. Res. 2022, 23, 1–6. [Google Scholar] [CrossRef]
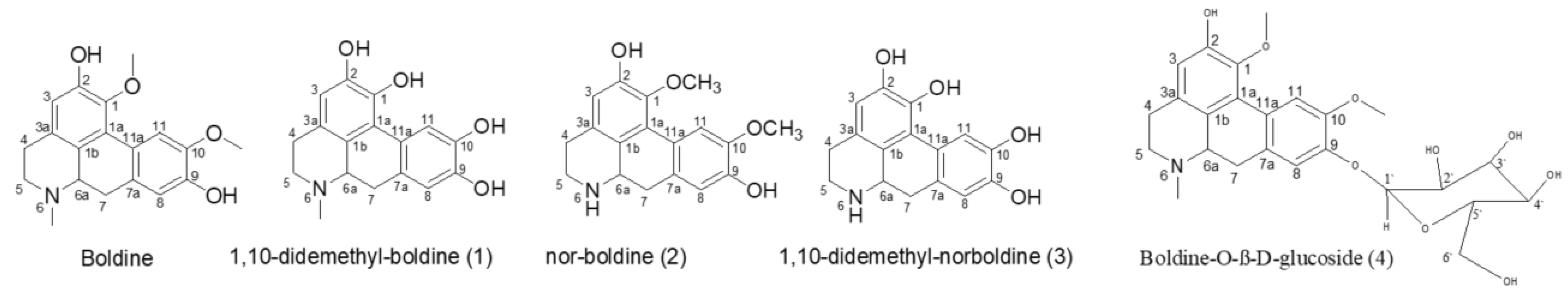
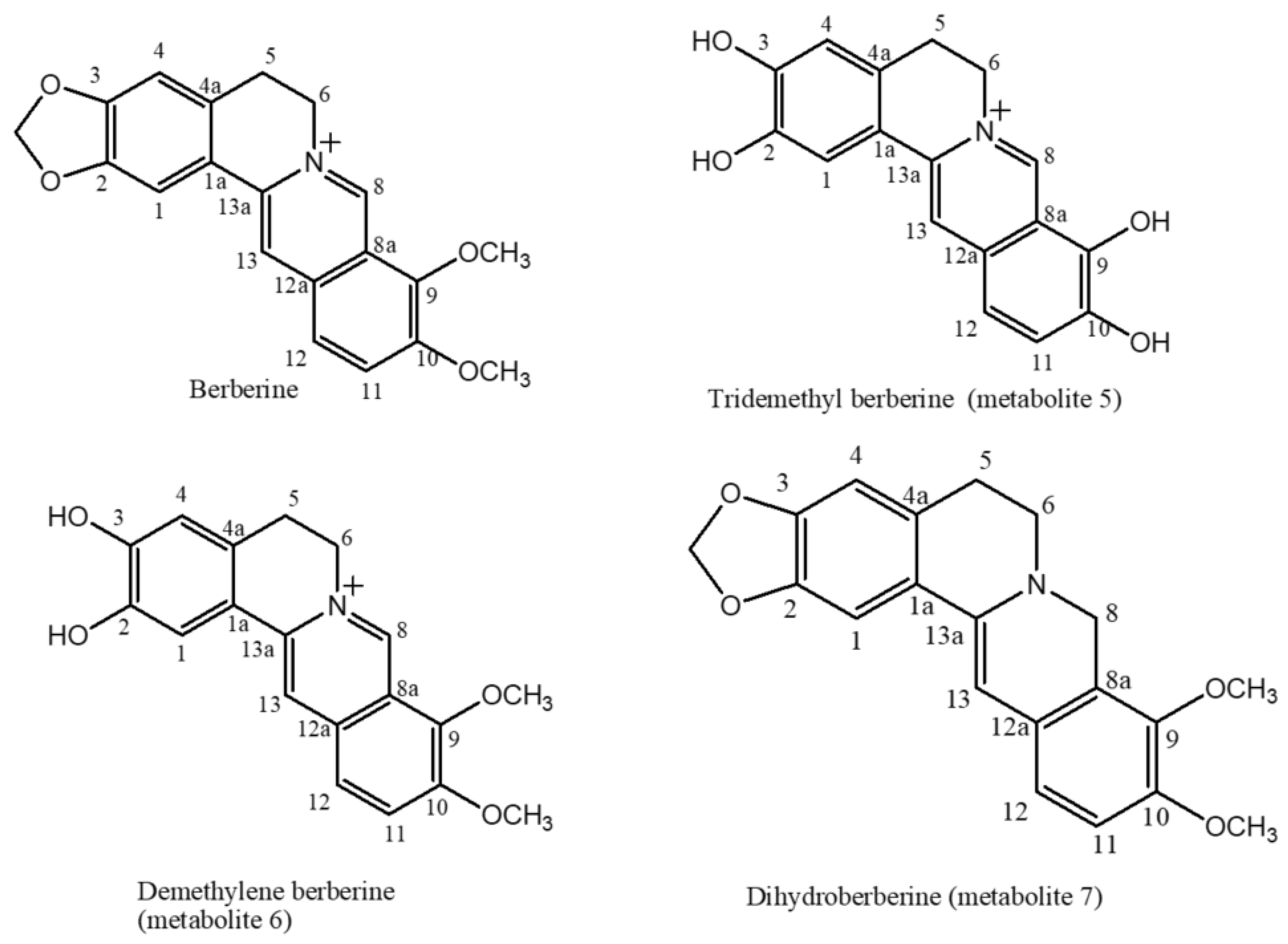
| Position | δH, Integration, Multiplicities (J in Hz) | δC, Multiplicities | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Boldine | Boldine Metabolite-1 | Boldine Metabolite-2 | Boldine Metabolite-3 | Boldine Metabolite-4 | Boldine | Boldine Metabolite-1 | Boldine Metabolite-2 | Boldine Metabolite-3 | Boldine Metabolite-4 | |
| 1 | --- | --- | --- | --- | --- | 142.6, C | 138.3, C | 143.3, C | 139.3, C | 141.8, C |
| 1a | --- | --- | --- | --- | --- | 122.8, C | 125.3, C | 125.3, C | 123.6, C | 126.1, C |
| 1b | --- | --- | --- | --- | --- | 126.2, C | 123.8, C | 126.2, C | 125.3, C | 125.0, C |
| 2 | --- | --- | --- | --- | --- | 149.1, C | 149.3, C | 149.0, C | 147.5, C | 148.3, C |
| 3 | 6.49, 1H, s | 6.52, 1H, s | 6.64, 1H, s | 6.48, 1H, s | 6.52, 1H, s | 114.1, CH | 114.6, CH | 115.0, CH | 112.2, CH | 114.2, CH |
| 3a | --- | --- | --- | --- | --- | 128.8, C | 128.2, C | 128.2, C | 128.6, C | 130.0, C |
| 4 | 2.24, 2H, m | 2.52, 2H, br | 2.52, 2H, br | 2.77, 2H, br | 2.51, 2H, m | 28.5, CH2 | 28.1, CH2 | 28.9, CH2 | 26.3, CH2 | 28.4, CH2 |
| 5 | 2.50, 2H, m | 2.97, 2H, br | 3.02, 2H, br | 2.94, 2H, br | 2.96, 2H, m | 52.8, CH2 | 53.4, CH2 | 42.4, CH2 | 45.1, CH2 | 52.4, CH2 |
| 6a | 2.73, 1H, dd (3.72, 13.8) | 2.66, 1H, m | 2.65, 1H, m | 2.90, 1H, m | 2.65, 1H, m | 50.3, CH | 60.8, CH | 52.7, CH | 55.3, CH | 61.3, CH |
| 7 | 2.83, 2H, m | 3.15, 2H, m | 3.20, 2H, m | 3.11, 2H, m | 3.20, 2H, m | 33.8, CH2 | 32.3, CH2 | 34.7, CH2 | 35.1, CH2 | 33.3, CH2 |
| 7a | --- | --- | --- | --- | --- | 129.6, C | 128.6, C | 130.6, C | 130.4, C | 130.6, C |
| 8 | 6.71, 1H, s | 6.68, 1H, s | 6.74, 1H, s | 6.68, 1H, s | 6.68, 1H, s | 115.2, CH | 115.4, CH | 114.9, CH | 115.7, CH | 118.4, CH |
| 9 | --- | --- | --- | --- | --- | 146.0, C | 144.5, C | 145.1, C | 147.3, C | 146.2, C |
| 10 | --- | --- | --- | --- | --- | 145.8, C | 142.5, C | 145.7, C | 140.2, C | 149.3, C |
| 11 | 7.85, 1H, s | 7.87, 1H, s | 7.89, 1H, s | 7.73, 1H, s | 7.87, 1H, s | 111.9, CH | 110.3, CH | 112.2, CH | 111.2, CH | 110.4, CH |
| 11a | --- | --- | --- | --- | --- | 125.6, C | 120.2, C | 123.2, C | 122.5, C | 127.3, C |
| 1-OCH3 | 3.55, 3H, s | --- | 3.58, 3H, s | --- | 3.56, 3H, s | 59.2, CH3 | --- | 58.4, CH3 | --- | 59.1, CH3 |
| 10-OCH3 | 3.77, 3H, s | --- | 3.76, 3H, s | --- | 3.77, 3H, s | 55.7, CH3 | --- | 55.3, CH3 | --- | 55.2, CH3 |
| N-CH3 | 2.38, 3H, s | 2.38, 3H, s | --- | --- | 2.37, 3H, s | 43.7, CH3 | 44.3, CH3 | --- | --- | 44.2, CH3 |
| Glc-1` | --- | --- | --- | --- | 5.05, 1H, d (7.2) | --- | --- | --- | --- | 101.2, CH |
| Glc-2` | --- | --- | --- | --- | 4.00–4.26, 6H, m | --- | --- | --- | --- | 77.2, CH |
| Glc-3` | --- | --- | --- | --- | --- | --- | --- | --- | 77.0, CH | |
| Glc-4` | --- | --- | --- | --- | --- | --- | --- | --- | 73.9, CH | |
| Glc-5` | --- | --- | --- | --- | --- | --- | --- | --- | 70.4, CH | |
| Glc-6` | --- | --- | --- | --- | --- | --- | --- | --- | 61.8, CH2 | |
| Position | δH, Integration, Multiplicities (J in Hz) | δC, Multiplicities | ||||||
|---|---|---|---|---|---|---|---|---|
| Berberine | Berberine Metabolite-5 | Berberine Metabolite-6 | Berberine Metabolite-7 | Berberine | Berberine Metabolite-5 | Berberine Metabolite-6 | Berberine Metabolite-7 | |
| 1 | 7.67, 1H, s | 7.82, 1H, s | 7.57, 1H, s | 6.73, 1H, s | 105.4, CH | 104.7, CH | 105.8, CH | 106.7, CH |
| 1a | --- | --- | --- | --- | 120.4, C | 122.5, C | 120.5, C | 122.3, C |
| 2 | --- | --- | --- | --- | 147.6, C | 148.2, C | 146.9, C | 145.9, C |
| 3 | --- | --- | --- | --- | 149.7, C | 150.3, C | 149.5, C | 148.3, C |
| 4 | 6.97, 1H, s | 7.07, 1H, s | 6.87, 1H, s | 6.58, 1H, s | 108.4, CH | 108.0, CH | 109.0, CH | 110.5, CH |
| 4a | --- | --- | --- | --- | 130.6, C | 131.7, C | 128.0, C | 129.3, C |
| 5 | 3.27, 2 H, t (4) | 3.21, 2H, t (4) | 3.12, 2H, t (4) | 2.91, 2H, t (4) | 26.4, CH2 | 27.0, CH2 | 25.9, CH2 | 28.4, CH2 |
| 6 | 4.94, 2 H, t (4) | 4.94, 2H, t (4) | 4.89, 2H, t (4) | 3.21, 2H, t (4) | 55.2, CH2 | 54.1, CH2 | 55.0, CH2 | 53.1, CH2 |
| 8 | 9.77, 1H, s | 9.87, 1H, s | 9.82, 1H, s | 4.30, 2H, s | 145.4, CH | 143.9, CH | 144.3, CH | 51.4, CH2 |
| 8a | --- | --- | --- | --- | 121.4, C | 122.4, C | 120.9, C | 120.7, C |
| 9 | --- | --- | --- | --- | 143.6, C | 145.4, C | 144.3, C | 145.2, C |
| 10 | --- | --- | --- | --- | 150.4, C | 151.4, C | 150.3, C | 149.2, C |
| 11 | 8.12, 1H, d (9) | 8.33, 1H, d (8) | 8.18, 1H, d (8) | 7.18, 1H, d (8) | 126.7, CH | 125.9, CH | 126.2, CH | 125.0, CH |
| 12 | 8.01, 1H, d (9) | 7.92, 1H, d (8) | 7.99, 1H, d (8) | 7.22, 1H, d (8) | 123.5, CH | 123.2, CH | 123.7, CH | 121.5, CH |
| 12a | --- | --- | --- | --- | 132.9, C | 133.0, C | 133.0, C | 130.5, C |
| 13 | 8.71, 1H, s | 8.88, 1H, s | 8.75, 1H, s | 5.93, 1H, s | 120.2, CH | 121.4, CH | 119.8, CH | 100.8, CH |
| 13a | --- | --- | --- | --- | 137.4, C | 136.8, C | 136.7, C | 138.4, C |
| O-CH2-O | 6.11, 2H, s | --- | --- | 5.90, 2H, s | 102.1, CH2 | --- | --- | 101.2, CH2 |
| O-9-CH3 | 4.21, 3H, s | --- | 4.10, 3H, s | 4.20, 3H, s | 62.0, CH3 | --- | 61.4, CH3 | 61.3, CH3 |
| O-10-CH3 | 4.11, 3H, s | --- | 4.07, 3H, s | 3.80, 3H, s | 57.1, CH3 | --- | 56.5, CH3 | 56.2, CH3 |
| Compound | TERT | Compound | PTP-1B | ||
|---|---|---|---|---|---|
| MolDock Score | Interacting Amino Acid | MolDock Score | Interacting Amino Acid | ||
| Boldine | −120.14 | Arg194, Asp343, Tyr256 | Berberine | −123.57 | Cys215, Ser216, Gly220, Arg221 |
| Boldine metabolite 1 | −112.52 | Asp343, Gly309 | Berberine metabolite 5 | −115.91 | Cys215, Ser216, Arg221 |
| Boldine metabolite 2 | −135.54 | Asp343, Gly309 | Berberine metabolite 6 | −115.60 | Cys215, Ser216, Arg221 |
| Boldine metabolite 3 | −119.00 | Asp343, Gly309, Tyr256 | Berberine metabolite 7 | −106.05 | Cys215, Ile219, Gly220, Arg221 |
| Boldine metabolite 4 | −151.75 | Arg194, Asp254, Ala255, Lys372, Lys189 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eliwa, D.; Ibrahim, A.-R.S.; Kabbash, A.; El-Aasr, M.; Tomczyk, M.; Bin Jardan, Y.A.; Batiha, G.E.-S.; Ragab, A.E. Biotransformation of Modified Benzylisoquinoline Alkaloids: Boldine and Berberine and In Silico Molecular Docking Studies of Metabolites on Telomerase and Human Protein Tyrosine Phosphatase 1B. Pharmaceuticals 2022, 15, 1195. https://doi.org/10.3390/ph15101195
Eliwa D, Ibrahim A-RS, Kabbash A, El-Aasr M, Tomczyk M, Bin Jardan YA, Batiha GE-S, Ragab AE. Biotransformation of Modified Benzylisoquinoline Alkaloids: Boldine and Berberine and In Silico Molecular Docking Studies of Metabolites on Telomerase and Human Protein Tyrosine Phosphatase 1B. Pharmaceuticals. 2022; 15(10):1195. https://doi.org/10.3390/ph15101195
Chicago/Turabian StyleEliwa, Duaa, Abdel-Rahim S. Ibrahim, Amal Kabbash, Mona El-Aasr, Michał Tomczyk, Yousef A. Bin Jardan, Gaber El-Saber Batiha, and Amany E. Ragab. 2022. "Biotransformation of Modified Benzylisoquinoline Alkaloids: Boldine and Berberine and In Silico Molecular Docking Studies of Metabolites on Telomerase and Human Protein Tyrosine Phosphatase 1B" Pharmaceuticals 15, no. 10: 1195. https://doi.org/10.3390/ph15101195
APA StyleEliwa, D., Ibrahim, A.-R. S., Kabbash, A., El-Aasr, M., Tomczyk, M., Bin Jardan, Y. A., Batiha, G. E.-S., & Ragab, A. E. (2022). Biotransformation of Modified Benzylisoquinoline Alkaloids: Boldine and Berberine and In Silico Molecular Docking Studies of Metabolites on Telomerase and Human Protein Tyrosine Phosphatase 1B. Pharmaceuticals, 15(10), 1195. https://doi.org/10.3390/ph15101195












