Rational Design, Synthesis and Biological Evaluation of Novel Pyrazoline-Based Antiproliferative Agents in MCF-7 Cancer Cells
Abstract
:1. Introduction
Rationale of the Work
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
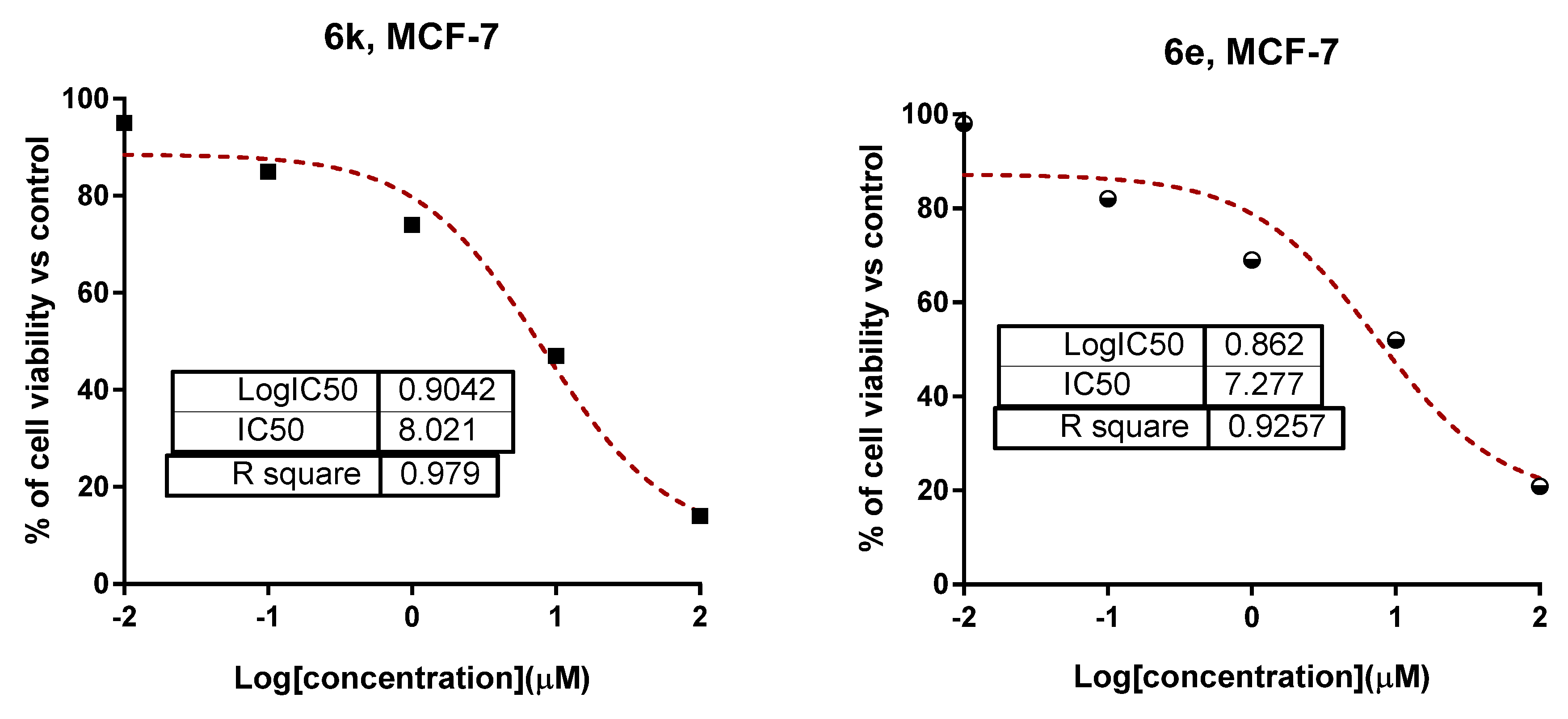
2.2.1. Cytotoxicity
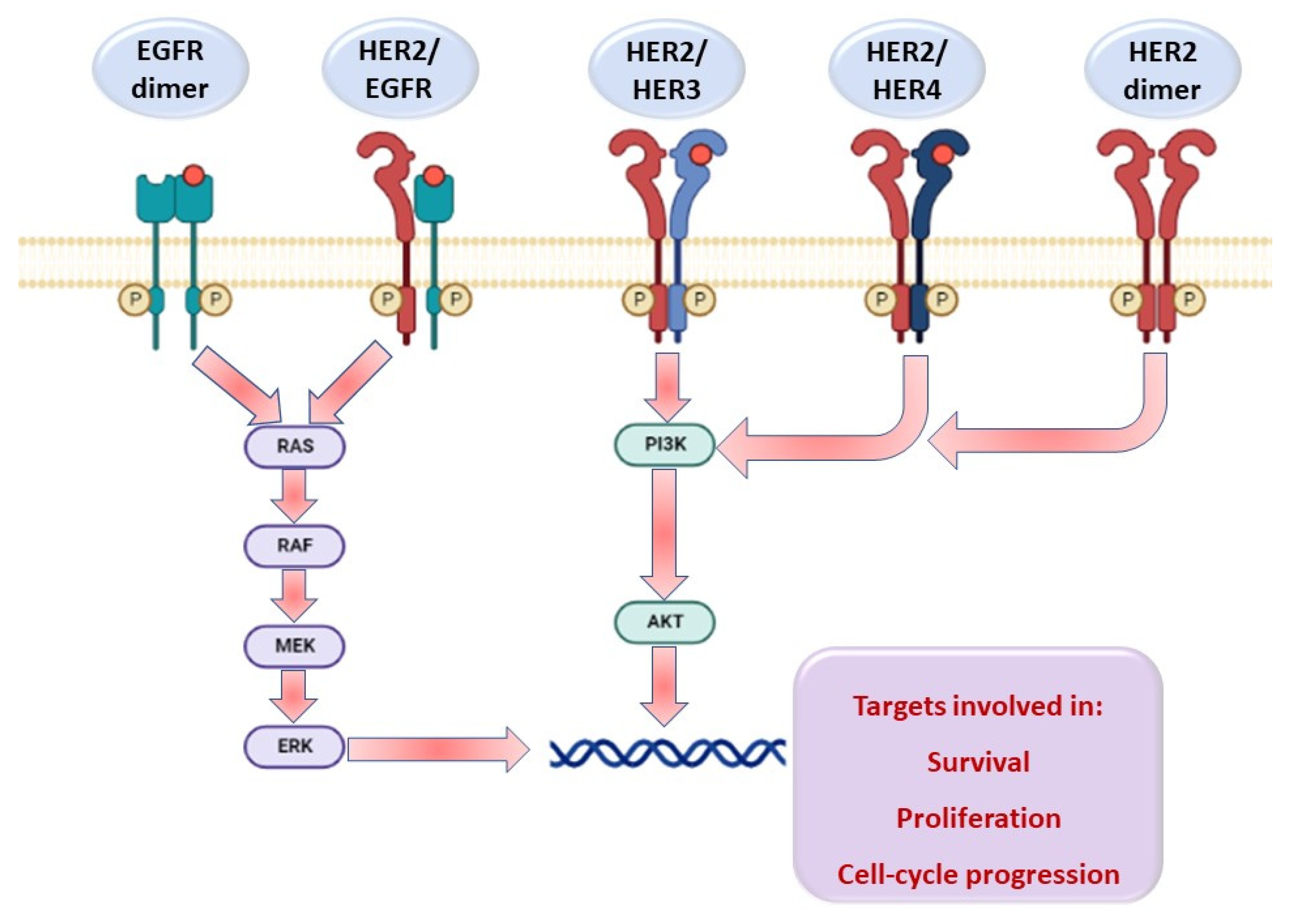
2.2.2. Potential EGFR/HER2 Kinase Inhibitory Assay
2.2.3. Apoptotic Investigation
Annexin V/PI Staining with Cell Cycle Analysis
RT-PCR
2.2.4. In Vivo Studies
2.3. Computational Studies
2.3.1. Molecular Docking
- (1)
- Compound 6e inside the binding site of the EGFR receptor (binding free energy = −13.05 kcal/mol) formed H bonds with Met793, Pro749, Arg841, Cys797, Gly796, and Leu792 at distances of 4.08, 2.69, 2.45, 2.03, 3.36 and 4.23 Å, respectively. Besides, π–H bonds with Lys745 and Val726 were formed. For HER2, compound 6e (binding free energy = −11.92 kcal/mol) showed H bonds with Cys805, Lys753, Thr862, Val734 and Asp863 at distance of 2.09, 2.31, 4.3, 3.99 and 3.32 Å, respectively. In addition, a π–H bond with Val734 was formed.
- (2)
- Compound 6k inside the binding site of EGFR receptor (binding free energy = −13.04 kcal/mol) achieved an H bond with Met793, Asp855, and Lys745 at a distance of 2.85, 3.05, 2.45 Å, respectively. Π–H bond with Val726, Leu718, Arg841, and Gly719 were formed. Furthermore, inside the binding site of HER2 receptor, compound 6k (binding free energy = −13.11 kcal/mol) established H bonds with Met801, Arg849, Phe864, Val734, and Asp863 at distance of 2.76, 2.13, 3.54, 4.26 and 3.1 Å, respectively. Besides π–H bonds with Cys805, Lys753 and Leu852 were noticed.
- (3)
- Compound 3c inside the binding site of EGFR receptor (binding free energy = −13.36 kcal/mol) achieved H bonds with Met793, Asp855, Lys745, and Gly796 at distance of 3.04, 2.99, 3.36, and 3.56 Å, in addition to the formation of π–H bonds with Leu718 and Phe856. Moreover, regarding the binding pocket of Her2 receptor (binding free energy = −11.3 kcal/mol), compound 3c formed H bonds with Lys753, Asp863, and Thr862 at a distance of 3.06, 2.77, and 4.23 Å, respectively and one π–H bond with Leu785.
- (4)
- For compound 9f inside the EGFR receptor binding site (binding free energy = −13.93 kcal/mol), H bonds with Met793, Asp855, Thr854, Lys745, Val726, and Met1002 were formed at distance of 3.05, 3.2, 2.18, 2.61, 3.55, and 3.8 Å, respectively, in addition to π–H bonds with Lys745 and Val726. On the other hand, compound 9f binding within the binding pocket of HER2 receptor (binding free energy = −11.85 kcal/mol) showed H bonds with Met801, Lys753, and Leu852 at distance of 3.01, 2.69, and 4.09 Å, respectively, besides one π–H bond with Val734.
2.3.2. In Silico Physicochemical Descriptors, Pharmacokinetic Properties and Bioactivity Prediction
2.3.3. Structure–Activity Relationship (SAR) Study
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of 3-(4-Aryl)-1-(3,4-dimethoxyphenyl)prop-2-en-1-one (2a–c)
3.1.2. Synthesis of 3-(3,4-Dimethoxyphenyl)-5-(4-aryl)-4,5-dihydro-1H-pyrazole-1-carbothioamide (3a–c)
Synthesis of 3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazole-1-carbothioamide (3a)
Synthesis of 3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazole-1-carbothioamide (3b)
Synthesis of 5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazole-1-carbothioamide (3c)
3.1.3. Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-(aryl)-4,5-dihydro-1H-pyrazol-1-yl)-4-(aryl1) Thiazole
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)-4-phenylthiazole (6a)
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)-4-(p-tolyl)thiazole (6b)
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)-4-(4-fluorophenyl)thiazole (6c)
Synthesis of 4-(4-Chlorophenyl)-2-(3-(3,4-dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)thiazole (6d)
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-phenylthiazole (6e)
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-(p-tolyl)thiazole (6f)
Synthesis of 2-(3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-(4-fluorophenyl)thiazole (6g)
Synthesis of 4-(4-Chlorophenyl)-2-(3-(3,4-dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole (6h)
Synthesis of 2-(5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-phenylthiazole (6i)
Synthesis of 2-(5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-(p-tolyl)thiazole (6j)
Synthesis of 2-(5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-(4-fluorophenyl)thiazole (6k)
Synthesis of 4-(4-Chlorophenyl)-2-(5-(4-chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)thiazole (6l)
Synthesis of 1-(2-(3-(3,4-Dimethoxyphenyl)-5-aryl-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazol-5-yl)ethan-1-one (9a–f)
Synthesis of 1-(2-(3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazol-5-yl)ethan-1-one (9a)
Synthesis of 1-(2-(3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazol-5-yl)ethan-1-one (9b)
Synthesis of 1-(2-(5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazol-5-yl)ethan-1-one (9c)
Synthesis of Ethyl 2-(3-(3,4-Dimethoxyphenyl)-5-phenyl-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazole-5-carboxylate (9d)
Synthesis of Ethyl 2-(3-(3,4-Dimethoxyphenyl)-5-(4-methoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazole-5-carboxylate (9e)
Synthesis of Ethyl 2-(5-(4-Chlorophenyl)-3-(3,4-dimethoxyphenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-methylthiazole-5-carboxylate (9f)
3.2. Biological Evaluations
3.2.1. Cytotoxicity
3.2.2. EGFR/HER2 Kinase Inhibitory Assay
3.2.3. Investigation of Apoptosis
Annexin V/PI Staining and Cell Cycle Analysis
Real-Time Polymerase Chain Reaction for the Selected Genes
3.2.4. In Vivo Study
Ethics Statement
Animal, Tumor Inoculation Experiment Design
3.3. Computational Studies
3.3.1. Molecular Docking
3.3.2. In Silico Physicochemical Descriptors, Pharmacokinetic Properties, and Bioactivity Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sidney, S.; Go, A.S.; Rana, J.S. Transition From Heart Disease to Cancer as the Leading Cause of Death in the United States. Ann. Intern. Med. 2019, 171, 225. [Google Scholar] [CrossRef] [PubMed]
- Cheung-Ong, K.; Giaever, G.; Nislow, C. Biology, DNA-damaging agents in cancer chemotherapy: Serendipity and chemical biology. Chem. Biol. 2013, 20, 648–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeVita, V.T.; Chu, E. A history of cancer chemotherapy. Cancer Res. 2008, 68, 8643–8653. [Google Scholar] [CrossRef] [Green Version]
- Farber, S.; Diamond, L.K.; Mercer, R.D.; Sylvester, R.F., Jr.; Wolff, J.A. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid (aminopterin). N. Engl. J. Med. 1948, 238, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Hurley, L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 188–200. [Google Scholar] [CrossRef]
- Bennasroune, A.; Gardin, A.; Aunis, D.; Crémel, G.; Hubert, P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit. Rev. Oncol. Hematol. 2004, 50, 23–38. [Google Scholar] [CrossRef]
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Exp. Rev. Anticancer Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef]
- Bononi, A.; Agnoletto, C.; De Marchi, E.; Marchi, S.; Patergnani, S.; Bonora, M.; Giorgi, C.; Missiroli, S.; Poletti, F.; Rimessi, A. Protein kinases and phosphatases in the control of cell fate. Enzyme Res. 2011, 2011, 329098. [Google Scholar] [CrossRef]
- Gschwind, A.; Fischer, O.M.; Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Parang, K.; Sun, G. Protein kinase inhibitors in drug discovery. Pharm. Sci. Encycl. Drug Discov. Dev. Manuf. 2010, 1–67. [Google Scholar]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [Green Version]
- Roskoski, R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Hu, S.; Sun, Y.; Meng, Y.; Wang, X.; Yang, W.; Fu, W.; Guo, H.; Qian, W.; Hou, S.; Li, B.; et al. Molecular architecture of the ErbB2 extracellular domain homodimer. Oncotarget 2015, 6, 1695–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkhipov, A.; Shan, Y.; Das, R.; Endres, N.F.; Eastwood, M.P.; Wemmer, D.E.; Kuriyan, J.; Shaw, D.E. Architecture and Membrane Interactions of the EGF Receptor. Cell 2013, 152, 557–569. [Google Scholar] [CrossRef] [Green Version]
- Rait, A.S.; Pirollo, K.F.; Rait, V.; Krygier, J.E.; Xiang, L.; Chang, E.H. Inhibitory effects of the combination of HER-2 antisense oligonucleotide and chemotherapeutic agents used for the treatment of human breast cancer. Cancer Gene Ther. 2001, 8, 728–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Othman, I.M.; Alamshany, Z.M.; Tashkandi, N.Y.; Gad-Elkareem, M.A.; Abd El-Karim, S.S.; Nossier, E.S.J.R.A. Synthesis and biological evaluation of new derivatives of thieno-thiazole and dihydrothiazolo-thiazole scaffolds integrated with a pyrazoline nucleus as anticancer and multi-targeting kinase inhibitors. RSC Adv. 2022, 12, 561–577. [Google Scholar] [CrossRef]
- Hennipman, A.; Van Oirschot, B.; Smits, J.; Rijksen, G.; Staal, G. Tyrosine kinase activity in breast cancer, benign breast disease, and normal breast tissue. Cancer Res. 1989, 49, 516–521. [Google Scholar]
- Tai, W.; Mahato, R.; Cheng, K.J. The role of HER2 in cancer therapy and targeted drug delivery. J. Control. Release 2010, 146, 264–275. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, K.A.; Wersig, T.; Slynko, I.; Totzke, F.; Schächtele, C.; Oelze, M.; Sippl, W.; Ritter, C.; Hilgeroth, A. Novel inhibitors of breast cancer relevant kinases Brk and HER2. MedChemComm 2014, 5, 659–664. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef]
- Li, J.Y.; Perry, S.R.; Muniz-Medina, V.; Wang, X.; Wetzel, L.K.; Rebelatto, M.C.; Hinrichs, M.J.M.; Bezabeh, B.Z.; Fleming, R.L.; Dimasi, N. A biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell 2016, 29, 117–129. [Google Scholar] [CrossRef] [Green Version]
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- Awadallah, F.M.; Piazza, G.A.; Gary, B.D.; Keeton, A.B.; Canzoneri, J.C. Synthesis of some dihydropyrimidine-based compounds bearing pyrazoline moiety and evaluation of their antiproliferative activity. Eur. J. Med. Chem. 2013, 70, 273–279. [Google Scholar] [CrossRef] [Green Version]
- George, R.F.; Fouad, M.A.; Gomaa, I.E. Synthesis and cytotoxic activities of some pyrazoline derivatives bearing phenyl pyridazine core as new apoptosis inducers. Eur. J. Med. Chem. 2016, 112, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Matiadis, D.; Sagnou, M.J. Pyrazoline hybrids as promising anticancer agents: An up-to-date overview. Int. J. Mol. Sci. 2020, 21, 5507. [Google Scholar] [CrossRef] [PubMed]
- Varghese, B.; Al-Busafi, S.N.; Suliman, F.O.; Al-Kindy, S.M. Unveiling a versatile heterocycle: Pyrazoline—A review. RSC Adv. 2017, 7, 46999–47016. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-J.; Lee, K.; Park, S.-J.; Ahn, B.; Lee, J.-C.; Cho, H.; Lee, K.-I.; Letters, M.C. Identification of antitumor activity of pyrazole oxime ethers. Bioorg. Med. Chem. Lett. 2005, 15, 3307–3312. [Google Scholar] [CrossRef] [PubMed]
- Hayat, F.; Salahuddin, A.; Umar, S.; Azam, A. Synthesis, characterization, antiamoebic activity and cytotoxicity of novel series of pyrazoline derivatives bearing quinoline tail. Eur. J. Med. Chem. 2010, 45, 4669–4675. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Arif, R.; Khan, F.I.; Maurya, V.; Singh, R.; Faizan, M.I.; Yasmeen, S.; Dar, S.H.; Alam, R.; Sahu, A. Pyrazoline analogs as potential anticancer agents and their apoptosis, molecular docking, MD simulation, DNA binding and antioxidant studies. Bioorg. Chem. 2021, 108, 104665. [Google Scholar] [CrossRef]
- Singh, P.; Singh, J.; Pant, G.J.; Rawat, M.S. 2-Pyrazolines as biologically active and fluorescent agents, an overview. Anti-Cancer Agents Med. Chem. 2018, 18, 1366–1385. [Google Scholar] [CrossRef] [PubMed]
- Alex, J.M.; Kumar, R.J. Chemistry, m. 4, 5-Dihydro-1 H-pyrazole: An indispensable scaffold. J. Enzyme Inhibit. Med. Chem. 2014, 29, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, M.R.; Mayhoub, A.S.; Farag, A.M. Recent advances in the therapeutic applications of pyrazolines. Exp. Opin. Ther. Pat. 2012, 22, 253–291. [Google Scholar] [CrossRef]
- Luan, S.; Zhong, H.; Zhao, X.; Yang, J.; Jing, Y.; Liu, D.; Zhao, L. Synthesis, anticancer evaluation and pharmacokinetic study of novel 10-O-phenyl ethers of dihydroartemisinin. Eur. J. Med. Chem. 2017, 141, 584–595. [Google Scholar] [CrossRef]
- Tilekar, K.; Upadhyay, N.; Meyer-Almes, F.J.; Loiodice, F.; Anisimova, N.Y.; Spirina, T.S.; Sokolova, D.V.; Smirnova, G.B.; Choe, J.Y.; Pokrovsky, V.S. Synthesis and Biological Evaluation of Pyrazoline and Pyrrolidine-2, 5-dione Hybrids as Potential Antitumor Agents. Chem. Med. Chem. 2020, 15, 1813–1825. [Google Scholar] [CrossRef]
- Amin, K.M.; Eissa, A.A.; Abou-Seri, S.M.; Awadallah, F.M.; Hassan, G.S. Synthesis and biological evaluation of novel coumarin–pyrazoline hybrids endowed with phenylsulfonyl moiety as antitumor agents. Eur. J. Med. Chem. 2013, 60, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Nepali, K.; Sharma, S.; Sharma, M.; Bedi, P.; Dhar, K.J.E. Rational approaches, design strategies, structure activity relationship and mechanistic insights for anticancer hybrids. Eur. J. Med. Chem. 2014, 77, 422–487. [Google Scholar] [CrossRef] [PubMed]
- Stephen, H.; Weizmann, C. Transactions, CI.—Derivatives of 3: 4-dimethoxyacetophenone and 4: 5-dimethoxy-o-tolyl methyl ketone, and the synthesis of phenylglyoxalines containing substituents in the benzene ring. J. Chem. Soc. Trans. 1914, 105, 1046–1057. [Google Scholar]
- Hay, M.P.; Turcotte, S.; Flanagan, J.U.; Bonnet, M.; Chan, D.A.; Sutphin, P.D.; Nguyen, P.; Giaccia, A.J.; Denny, W.A. 4-Pyridylanilinothiazoles that selectively target von Hippel− Lindau deficient renal cell carcinoma cells by inducing autophagic cell death. J. Med. Chem. 2010, 53, 787–797. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Inza, S.; Aydillo, C.; Sanmartín, C.; Plano, D. Thiazole moiety: An interesting scaffold for developing new antitumoral compounds. In Heterocycles-Synthesis and Biological Activities; IntechOpen: London, UK, 2019. [Google Scholar]
- Sharma, P.C.; Bansal, K.K.; Sharma, A.; Sharma, D.; Deep, A. Thiazole-containing compounds as therapeutic targets for cancer therapy. Eur. J. Med. Chem. 2020, 188, 112016. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Pepinosa, N.Y.; Cardona-Trujillo, M.C.; Garzon-Castano, S.C.; Veloza, L.A.; Sepúlveda-Arias, J.C.J.B. Pharmacotherapy, Antiproliferative activity of thiazole and oxazole derivatives: A systematic review of in vitro and in vivo studies. Biomed. Pharmacother. 2021, 138, 111495. [Google Scholar] [CrossRef] [PubMed]
- Merino, P.; Tejero, T.; Unzurrunzaga, F.J.; Franco, S.; Chiacchio, U.; Saita, M.G.; Iannazzo, D.; Piperno, A.; Romeo, G.J.T.A. An efficient approach to enantiomeric isoxazolidinyl analogues of tiazofurin based on nitrone cycloadditions. Tetrahedron Asymm. 2005, 16, 3865–3876. [Google Scholar] [CrossRef]
- Jain, S.; Pattnaik, S.; Pathak, K.; Kumar, S.; Pathak, D.; Jain, S.; Vaidya, A.J.M. Anticancer potential of thiazole derivatives: A retrospective review. Mini Rev. Med. Chem. 2018, 18, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.; Manzione, L.J.A.o.O. Dasatinib: A new step in molecular target therapy. Ann. Oncol. 2007, 18, vi42–vi46. [Google Scholar] [CrossRef]
- Donarska, B.; Świtalska, M.; Płaziński, W.; Wietrzyk, J.; Łączkowski, K.Z.J.B.C. Effect of the dichloro-substitution on antiproliferative activity of phthalimide-thiazole derivatives. Rational design, synthesis, elastase, caspase 3/7, and EGFR tyrosine kinase activity and molecular modeling study. Bioorg. Chem. 2021, 110, 104819. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, S.A.; Taher, E.S.; Lan, P.; Asaad, G.F.; Gomaa, H.A.; El-Koussi, N.A.; Youssif, B.G.J.B.C. Design, synthesis, and biological evaluation of new pyrimidine-5-carbonitrile derivatives bearing 1, 3-thiazole moiety as novel anti-inflammatory EGFR inhibitors with cardiac safety profile. Bioorg. Chem. 2021, 111, 104890. [Google Scholar] [CrossRef] [PubMed]
- Levantini, E.; Maroni, G.; Del Re, M.; Tenen, D.G. EGFR signaling pathway as therapeutic target in human cancers. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Qiu, K.-M.; Wang, H.-H.; Wang, L.-M.; Luo, Y.; Yang, X.-H.; Wang, X.-M.; Zhu, H.-L. Design, synthesis and biological evaluation of pyrazolyl-thiazolinone derivatives as potential EGFR and HER-2 kinase inhibitors. Bioorg. Med. Chem. 2012, 20, 2010–2018. [Google Scholar] [CrossRef] [PubMed]
- Gediya, L.K.; Njar, V.C. Promise and challenges in drug discovery and development of hybrid anticancer drugs. Exp. Opin. Drug Discov. 2009, 4, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, N.; Tilekar, K.; Loiodice, F.; Anisimova, N.Y.; Spirina, T.S.; Sokolova, D.V.; Smirnova, G.B.; Choe, J.-Y.; Meyer-Almes, F.-J.; Pokrovsky, V.S.J.B.C. Pharmacophore hybridization approach to discover novel pyrazoline-based hydantoin analogs with anti-tumor efficacy. Bioorg. Chem. 2021, 107, 104527. [Google Scholar] [CrossRef]
- Dhar, D.N. The Chemistry of Chalcones and Related Compounds; John Wiley & Sons: Hoboken, NJ, USA, 1981. [Google Scholar]
- Batt, D.G.; Goodman, R.; Jones, D.G.; Kerr, J.S.; Mantegna, L.R.; McAllister, C.; Newton, R.C.; Nurnberg, S.; Welch, P.K.; Covington, M.B. 2′-Substituted chalcone derivatives as inhibitors of interleukin-1 biosynthesis. J. Med. Chem. 1993, 36, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, M.; Sigler, S.; Racheed, N.A.; Hefnawy, A.; Fathalla, R.K.; Hammam, M.A.; Maher, A.; Maxuitenko, Y.; Keeton, A.B.; Hartmann, R.W.; et al. From Celecoxib to a Novel Class of Phosphodiesterase 5 Inhibitors: Trisubstituted Pyrazolines as Novel Phosphodiesterase 5 Inhibitors with Extremely High Potency and Phosphodiesterase Isozyme Selectivity. J. Med. Chem. 2021, 64, 4462–4477. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, L.-R.; Liu, L.-L.; Wang, W.; Man, R.-J.; Zheng, D.-J.; Deng, Y.-S.; Yang, Y.-S.; Xu, C.; Zhu, H.-L.; et al. A novel series of benzothiazepine derivatives as tubulin polymerization inhibitors with anti-tumor potency. Bioorg. Chem. 2021, 108, 104585. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.D.; Kulshreshtha, M.; Kumar, Y.; Chawla, A.P.; Ved, A.; Shukla, S.K. Design, Synthesis, Characterization and In Silico Molecular Docking Studies and In Vivo Anti-inflammatory Activity of Pyrazoline Clubbed Thiazolinone Derivatives. Lett. Org. Chem. 2021, 18, 735–748. [Google Scholar] [CrossRef]
- Singh, A.; Singour, P.K. QSAR studies, synthesis and biological evaluation of pyrazole derivatives containing thiourea as tyrosine kinase inhibitors: An approach to design anticancer agents. Int. J. Pharm. Sci. Res. 2020, 11, 4388–4394. [Google Scholar]
- Doğan, İ.S.; Sarac, S.; Sari, S.; Kart, D.; Gökhan, Ş.E.; Vural, I.; Dalkara, S.J. New azole derivatives showing antimicrobial effects and their mechanism of antifungal activity by molecular modeling studies. Eur. J. Med. Chem. 2017, 130, 124–138. [Google Scholar] [CrossRef]
- Abdel-Wahab, B.F.; Abdel-Aziz, H.A.; Ahmed, E.M. Synthesis and antimicrobial evaluation of 1-(benzofuran-2-yl)-4-nitro-3-arylbutan-1-ones and 3-(benzofuran-2-yl)-4,5-dihydro-5-aryl-1-[4-(aryl)-1,3-thiazol-2-yl]-1H-pyrazoles. Eur. J. Med. Chem. 2009, 44, 2632–2635. [Google Scholar] [CrossRef]
- Perrone, M.G.; Santandrea, E.; Dell’Uomo, N.; Giannessi, F.; Milazzo, F.M.; Sciarroni, A.F.; Scilimati, A.; Tortorella, V. Synthesis and biological evaluation of new clofibrate analogues as potential PPARα agonists. Eur. J. Med. Chem. 2005, 40, 143–154. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Rizk, H.F. Synthesis and biological evaluation of thiazole derivatives. In Azoles-Synthesis, Properties, Applications and Perspectives; IntechOpen: London, UK, 2020. [Google Scholar]
- Nagy, M.I.; Darwish, K.M.; Kishk, S.M.; Tantawy, M.A.; Nasr, A.M.; Qushawy, M.; Swidan, S.A.; Mostafa, S.M.; Salama, I. Design, synthesis, anticancer activity, and solid lipid nanoparticle formulation of indole-and benzimidazole-based compounds as pro-apoptotic agents targeting bcl-2 protein. Pharmaceuticals 2021, 14, 113. [Google Scholar] [CrossRef]
- Boraei, A.T.; Eltamany, E.H.; Ali, I.A.; Gebriel, S.M.; Nafie, M.S.J. Synthesis of new substituted pyridine derivatives as potent anti-liver cancer agents through apoptosis induction: In vitro, in vivo, and in silico integrated approaches. Bioorg. Chem. 2021, 111, 104877. [Google Scholar] [CrossRef]
- SwissADME. Available online: http://www.swissadme.ch/ (accessed on 2 July 2022).
- Wood, E.R.; Truesdale, A.T.; McDonald, O.B.; Yuan, D.; Hassell, A.; Dickerson, S.H.; Ellis, B.; Pennisi, C.; Horne, E.; Lackey, K.J. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib) relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004, 64, 6652–6659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, T.; Seto, M.; Banno, H.; Kawakita, Y.; Oorui, M.; Taniguchi, T.; Ohta, Y.; Tamura, T.; Nakayama, A.; Miki, H. Design and synthesis of novel human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors bearing a pyrrolo [3, 2-d] pyrimidine scaffold. J. Med. Chem. 2011, 54, 8030–8050. [Google Scholar] [CrossRef] [PubMed]
- Mosdam, T.J. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxic assay. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar]
- Tantawy, E.S.; Amer, A.M.; Mohamed, E.K.; Abd Alla, M.M.; Nafie, M.S. Synthesis, characterization of some pyrazine derivatives as anti-cancer agents: In vitro and in Silico approaches. J. Mol. Struct. 2020, 1210, 128013. [Google Scholar] [CrossRef]
- Hisham, M.; Youssif, B.G.; Osman, E.E.A.; Hayallah, A.M.; Abdel-Aziz, M. Synthesis and biological evaluation of novel xanthine derivatives as potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019, 176, 117–128. [Google Scholar] [CrossRef]
- Nafie, M.S.; Arafa, K.; Sedky, N.K.; Alakhdar, A.A.; Arafa, R. Triaryl dicationic DNA minor-groove binders with antioxidant activity display cytotoxicity and induce apoptosis in breast cancer. Chem. Biol. Interact. 2020, 324, 109087. [Google Scholar] [CrossRef]
- Nafie, M.S.; Kishk, S.M.; Mahgoub, S.; Amer, A.M. Quinoline-based thiazolidinone derivatives as potent cytotoxic and apoptosis-inducing agents through EGFR inhibition. Chem. Biol. Drug Des. 2022, 99, 547–560. [Google Scholar] [CrossRef]
- Gad, E.M.; Nafie, M.S.; Eltamany, E.H.; Hammad, M.S.; Barakat, A.; Boraei, A. Discovery of new apoptosis-inducing agents for breast cancer based on ethyl 2-amino-4,5,6,7-tetra hydrobenzo [b] thiophene-3-carboxylate: Synthesis, in vitro, and in vivo activity evaluation. Molecules 2020, 25, 2523. [Google Scholar] [CrossRef]
- Nafie, M.S.; Mahgoub, S.; Amer, A. Antimicrobial and antiproliferative activities of novel synthesized 6-(quinolin-2-ylthio) pyridine derivatives with molecular docking study as multi-targeted JAK2/STAT3 inhibitors. Chem. Biol. Drug Des. 2021, 97, 553–564. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inform. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AutoDock Vina Documentation. Release 1.2.0, Center of Computational Structural Biology (CCSB). Available online: https://autodock-vina.readthedocs.io/en/latest/introduction.html (accessed on 10 April 2022).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrrint, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- PreADMET. Available online: https://preadmet.qsarhub.com/ (accessed on 4 July 2022).
- Openmolecules. Available online: https://openmolecules.org/propertyexplorer/applet.html (accessed on 5 July 2022).













| Compounds | IC50 ± SD * (µM) | Compounds | IC50 ± SD * (µM) | ||
|---|---|---|---|---|---|
| MCF-7 | MCF-10A | MCF-7 | MCF-10A | ||
| 3a | NA | 36.5 ± 1.15 | 6i | 45.36 ± 1.87 | 15.82 ± 1.08 |
| 3b | 23.6 ± 0.72 | 34.8 ± 1.35 | 6j | NA | ≥50 |
| 3c | 11.19 ± 0.54 | ≥50 | 6k | 8.02 ± 0.98 | ≥50 |
| 6a | NA | ≥50 | 6l | 17.58 ± 0.97 | 35.7 ± 1.85 |
| 6b | 13.36 ± 0.98 | ≥50 | 9a | 34.25 ± 1.68 | 18.9 ± 1.09 |
| 6c | 18.69 ± 0.84 | ≥50 | 9b | 31.69 ± 1.96 | 26.34 ± 1.07 |
| 6d | NA | 45.2 ± 1.24 | 9c | NA | ≥50 |
| 6e | 7.21 ± 0.48 | ≥50 | 9d | NA | 24.3 ± 1.39 |
| 6f | 11.05 ± 0.85 | 35.4 ± 1.84 | 9e | 18.95 ± 1.03 | 43.6 ± 1.85 |
| 6g | 15.36 ± 0.79 | 24.5 ± 1.01 | 9f | 8.35 ± 0.29 | ≥50 |
| 6h | 35.25 ± 1.27 | 26.5 ± 1.59 | Lapatinib | 7.45 | ≥50 |
| Compound | IC50 [µM] * | |
|---|---|---|
| EGFR Kinase | HER2 Kinase | |
| 3c | 0.067± 0.009 | 0.07 ± 0.018 |
| 6f | 0.23 ± 0.024 | 0.12 ± 0.025 |
| 6e | 0.009 ± 0.001 | 0.013 ± 0.001 |
| 6k | 0.014 ± 0.001 | 0.027 ± 0.002 |
| 9f | 0.051 ± 0.003 | 0.14 ± 0.006 |
| Lapatinib | 0.006 ± 0.001 | 0.017 ± 0.002 |
| Sample | Fold Change = 2−ΔΔCT | |||||
|---|---|---|---|---|---|---|
| Proapoptotic Gene | Anti-Apoptotic Gene | |||||
| P53 | Bax | Casp-3 | Casp-8 | Casp-9 | Bcl-2 | |
| 6k- treated | 3.91 ± 0.50 | 5.24 ± 0.4 | 5.79 ± 0.4 | 3.91 ± 0.42 | 6.07 ± 0.23 | 0.41 ± 0.09 |
| 6e- treated | 4.85 ± 0.4 | 6.39 ± 0.8 | 7.12 ± 0.53 | 1.71 ± 0.18 | 8.19 ± 0.32 | 0.22 ± 0.06 |
| Untreated | 1 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fakhry, M.M.; Mahmoud, K.; Nafie, M.S.; Noor, A.O.; Hareeri, R.H.; Salama, I.; Kishk, S.M. Rational Design, Synthesis and Biological Evaluation of Novel Pyrazoline-Based Antiproliferative Agents in MCF-7 Cancer Cells. Pharmaceuticals 2022, 15, 1245. https://doi.org/10.3390/ph15101245
Fakhry MM, Mahmoud K, Nafie MS, Noor AO, Hareeri RH, Salama I, Kishk SM. Rational Design, Synthesis and Biological Evaluation of Novel Pyrazoline-Based Antiproliferative Agents in MCF-7 Cancer Cells. Pharmaceuticals. 2022; 15(10):1245. https://doi.org/10.3390/ph15101245
Chicago/Turabian StyleFakhry, Mariam M., Kazem Mahmoud, Mohamed S. Nafie, Ahmad O. Noor, Rawan H. Hareeri, Ismail Salama, and Safaa M. Kishk. 2022. "Rational Design, Synthesis and Biological Evaluation of Novel Pyrazoline-Based Antiproliferative Agents in MCF-7 Cancer Cells" Pharmaceuticals 15, no. 10: 1245. https://doi.org/10.3390/ph15101245








