Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs
Abstract
1. Introduction
2. Results
2.1. Chemical Assessment
2.1.1. Total Carbon (TC), Total Nitrogen (TN), Total Hydrogen (TH) and Total Sulphur (TS)
2.1.2. Protein, Carbohydrates, Lipids, Inorganic Compounds and Moisture
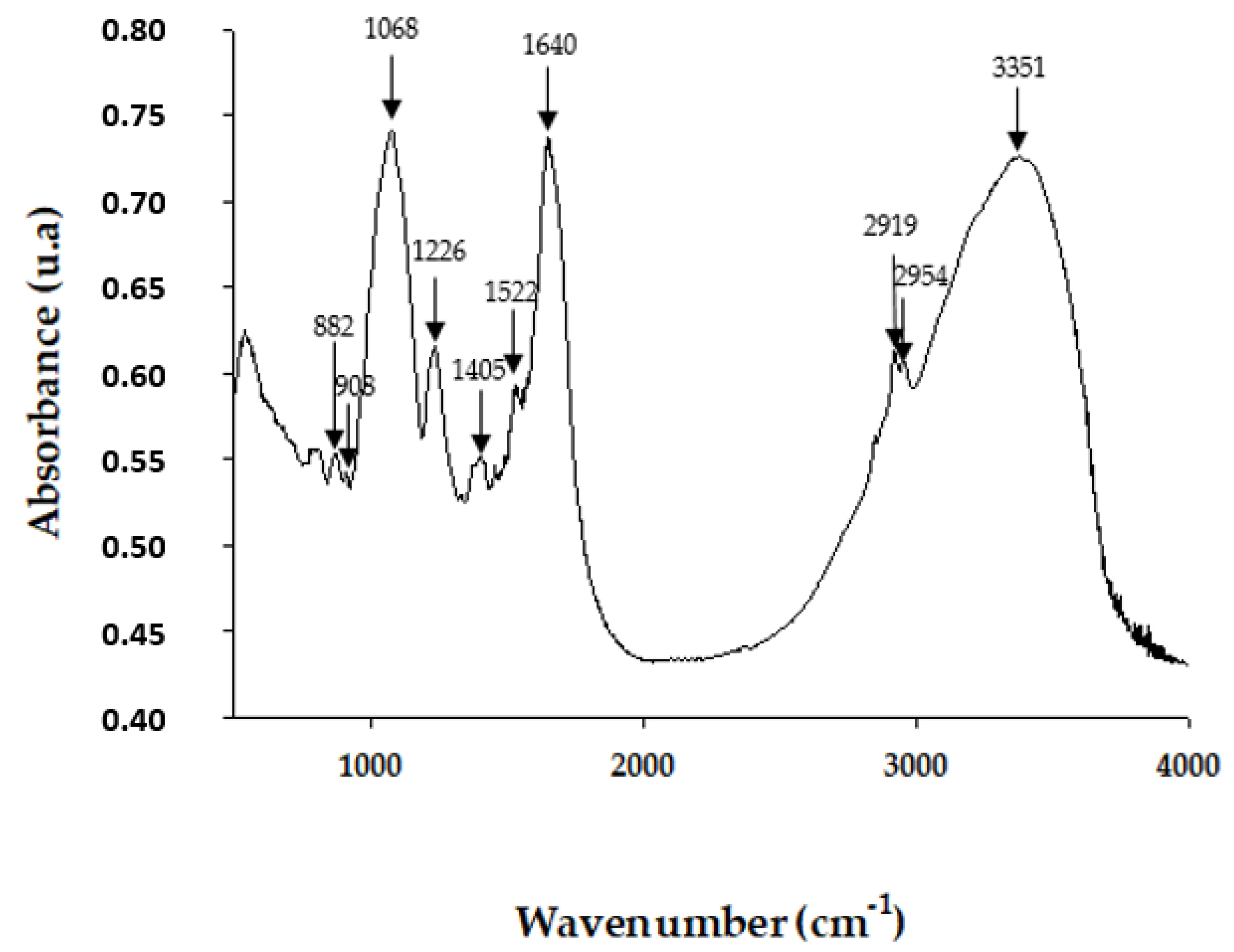
2.1.3. Fourier Transform Infrared Spectroscopy (FT-IR)
2.1.4. Gas Chromatography–Mass Spectrometry (GC-MS)
2.2. Biological Assessment
2.2.1. Antioxidant Activity (ABTS Method)
2.2.2. Antioxidant Activity (DPPH Method)
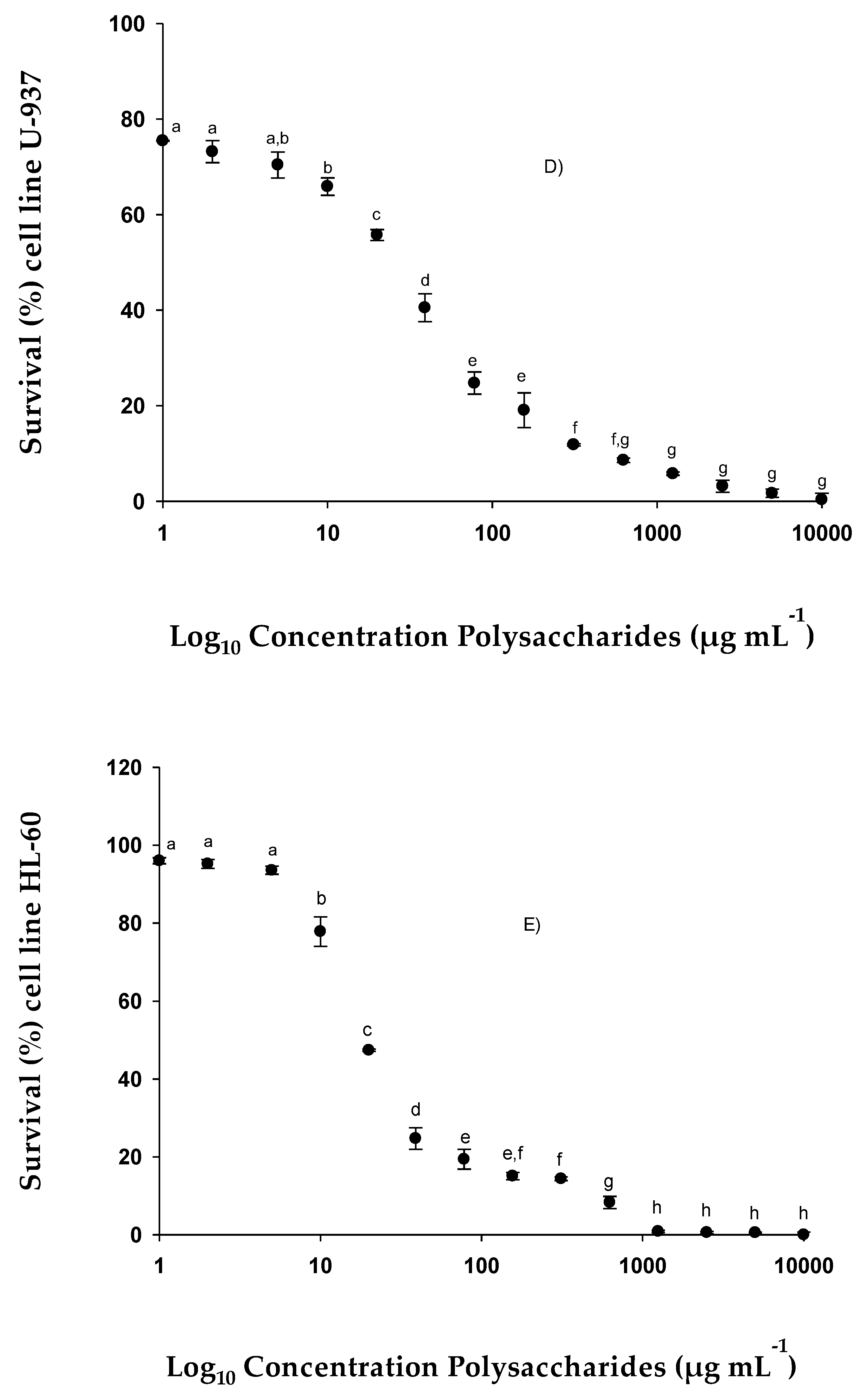
2.2.3. Cell Viability of Lines HTC-116, MCF-7, U-937, HL-60 and NCl-H460
2.2.4. Cell Viability of Line HFG-1 more and less Proliferating
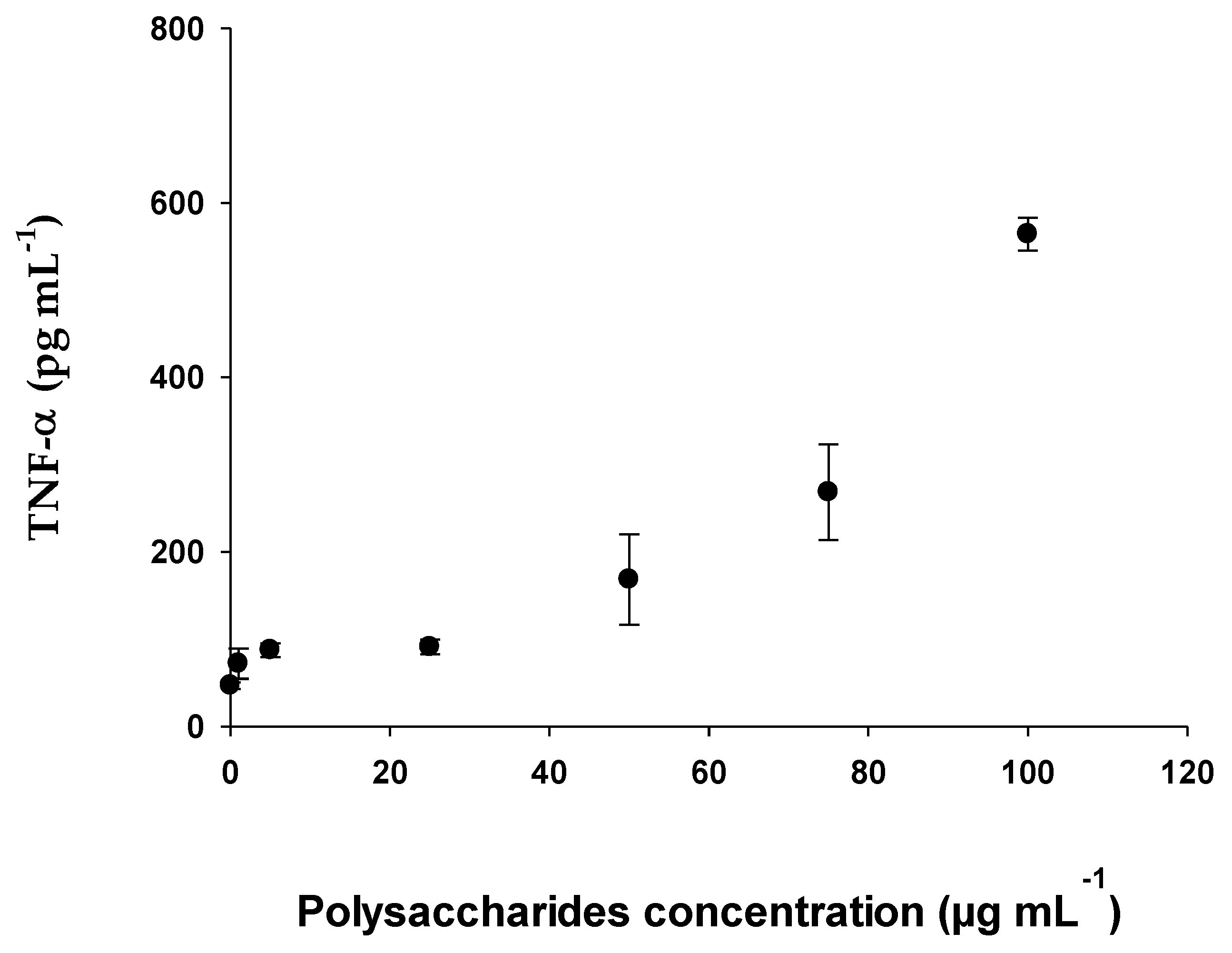
2.2.5. Determination of Cytokines (IL-6 and TNF-α) with RAW 264.7 Cell Line
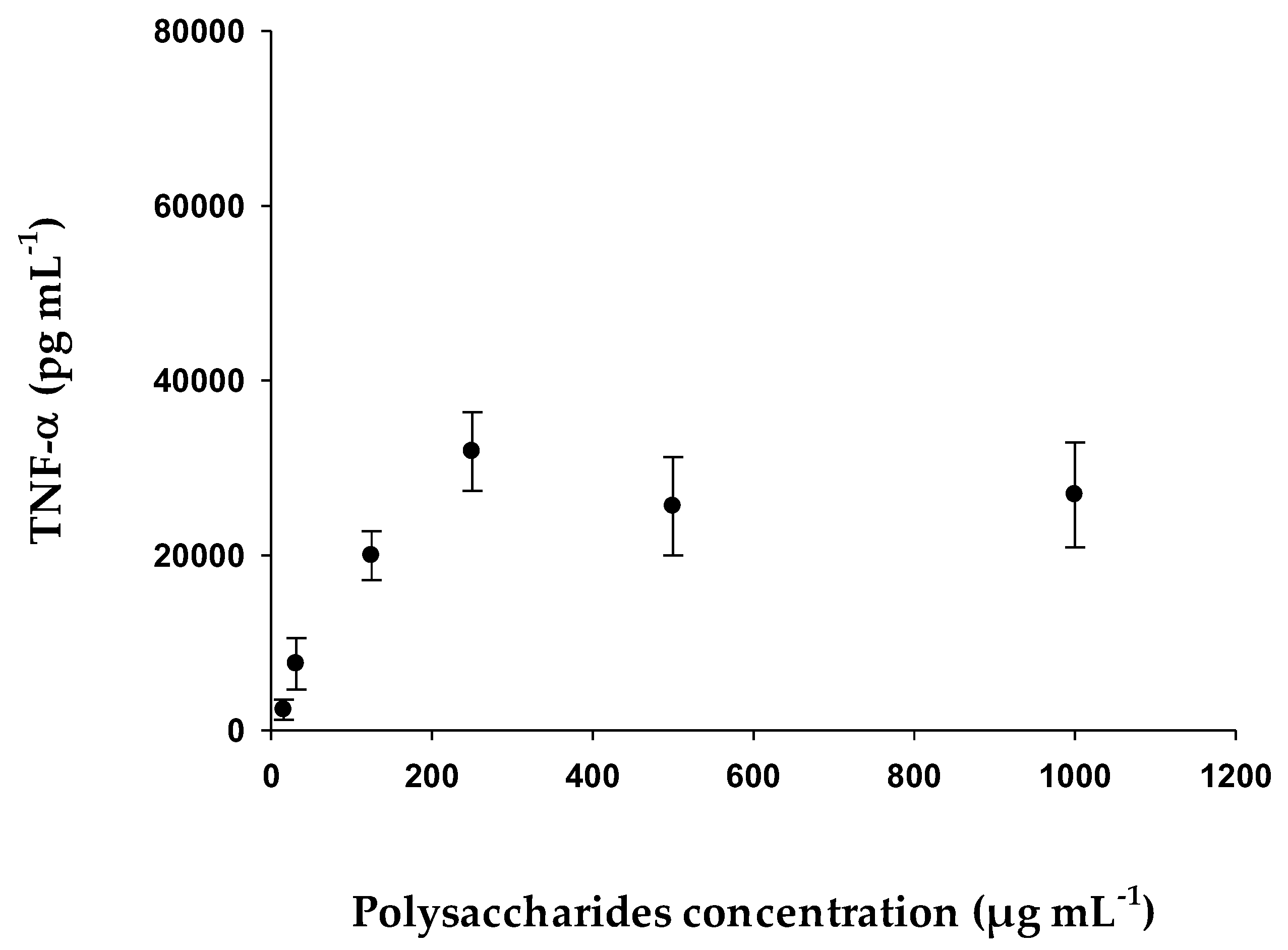
2.2.6. Determination of TNF-α with Human THP-1 Cell Line
2.2.7. Proteomic Analysis in HGF-1 Cells
2.3. STRING Analysis of Protein Networks
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Total Carbon (C), Hydrogen (H), Nitrogen (N) and Sulphur (S)
4.3. Protein Content
4.4. Determination of Total Carbohydrates
4.5. Determination of Lipids
4.6. Determination of Inorganic Compounds
4.7. Extraction of Polysaccharides
4.8. Fourier Transform Infrared Spectroscopy (FT-IR)
4.9. Gas Chromatography–Mass Spectrometry (GC-MS)
4.9.1. Derivatization of Polysaccharides
4.9.2. Gas Chromatography/Mass Spectrometry (GC-MS) Analysis
4.10. Antioxidant Activity (ABTS Method) in Polysaccharides and Biomass
4.11. Antioxidant Activity (DPPH Method) in Biomass
4.12. Lipopolysaccharides (LPS) Contamination Assay
4.13. Cell Culture
4.14. MTT Assay in Tumoral Cell Lines
4.15. Cytotoxicity Assay MTT with Healthy Cell Line (HGF-1)
4.16. Determination of Cytokines with RAW 264.7 Cell Line
4.17. Determination of Cytokines with Human THP-1 Cell Line
4.18. Proteomics Analysis
4.18.1. Cell Treatment and Protein Extraction
4.18.2. In-Solution Tryptic Digestion and 2-Plex Tandem Mass Tag (TMT) Labeling
4.18.3. Liquid Chromatography High-Resolution Mass Spectrometry
4.18.4. Data Analysis
4.19. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irigoien, X.; Huisman, J.; Harris, R. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature 2004, 429, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Talero, E.; García-Mauriño, S.; Ávila-Román, J.; Rodríguez-Luna, A.; Alcaide, A.; Motilva, V. Bioactive compounds isolated from microalgae in chronic inflammation and cancer. Mar. Drugs 2015, 13, 6152–6209. [Google Scholar] [CrossRef] [PubMed]
- Mohd Syahril, M.; Roshani, O.; Nur Hasyimah, R.; Mohamad Hafiz, M.; Sharida, M.; Ahmed, H. Screening of anticancer activities of crude extracts of unicellular green algae (Chlorella vulgaris) and filamentous blue green algae (Spirulina platensis) on selected cancer cell lines. J. Acad. 2012, 2, 38–42. [Google Scholar]
- Rupérez, P.; Ahrazem, O.; Leal, J.A. Potential Antioxidant Capacity of Sulfated Polysaccharides from the Edible Marine Brown Seaweed Fucus vesiculosus. J. Agric. Food Chem. 2002, 50, 840–845. [Google Scholar] [CrossRef]
- Jiao, G.; Yu, G.; Zhang, J.; Ewart, H.S. Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar. Drugs 2011, 9, 196–233. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, S.; Ermakova, S.; Zvyagintseva, T.; Stonik, V. Anticancer and Cancer Preventive Properties of Marine Polysaccharides: Some Results and Prospects. Mar. Drugs 2013, 11, 4876–4901. [Google Scholar] [CrossRef] [PubMed]
- Osafune, T.; Sumida, S.; Ehara, T.; Ueno, N.; Hase, E.; Schiff, J.A. Lipid (wax) and Paramylum as Sources of Carbon and Energy for the Early Development of Proplastids in Dark-Grown Euglena gracilis Cells Transferred to an Inorganic Medium. J. Electron. Microsc. 1990, 39, 372–381. [Google Scholar]
- Takeyama, H.; Kanamaru, A.; Yoshino, Y.; Kakuta, H.; Kawamura, Y.; Matsunaga, T. Production of antioxidant vitamins, β-carotene, vitamin C, and vitamin E, by two-step culture of Euglena gracilis Z. Biotechnol. Bioeng. 1997, 53, 185–190. [Google Scholar] [CrossRef]
- Ogbonna, J.C.; Tomiyamal, S.; Tanaka, H. Heterotrophic cultivation of Euglena gracilis Z for efficient production of α-tocopherol. J. Appl. Phycol. 1998, 10, 67–74. [Google Scholar] [CrossRef]
- Fujita, T.; Aoyagi, H.; Ogbonna, J.C.; Tanaka, H. Effect of mixed organic substrate on α-tocopherol production by Euglena gracilis in photoheterotrophic culture. Appl. Microbiol. Biotechnol. 2008, 79, 371–378. [Google Scholar] [CrossRef]
- Barsanti, L.; Vismara, R.; Passarelli, V.; Gualtieri, P. Paramylon (β-1,3-glucan) content in wild type and WZSL mutant of Euglena gracilis. Effects of growth conditions. J. Appl. Phycol. 2001, 13, 59–65. [Google Scholar] [CrossRef]
- Fruehauf, J.P.; Bonnard, G.D.; Herberman, R.B. The effect of lentinan on production of interleukin-1 by human monocytes. Immunopharmacology 1982, 5, 65–74. [Google Scholar] [CrossRef]
- Sugawara, I.; Lee, K.; Wong, M. Schizophyllan (SPG)-treated macrophages and anti-tumor activities against syngeneic and allogeneic tumor cells. Cancer Immunol. Immunother. 1984, 16, 137–144. [Google Scholar] [CrossRef]
- Fish, A.; April, S.I.; Skov, J.; Buchmann, K. Immunomodulatory effects of dietary β-1,3-glucan from Euglena gracilis in rainbow trout (Oncorhynchus mykiss) immersion vaccinated against Yersinia ruckeri. Fish Shellfish. Immunol. 2012, 33, 111–120. [Google Scholar] [CrossRef]
- Watanabe, T.; Shimada, R.; Matsuyama, A.; Yuasa, M.; Sawamura, H.; Yoshida, E.; Suzuki, K. Antitumor activity of the β-glucan paramylon from Euglena against preneoplastic colonic aberrant crypt foci in mice. Food Funct. 2013, 4, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Barsanti, L.; Passarelli, V.; Evangelista, V.; Frassanito, A.M.; Gualtieri, P. Chemistry, physico-chemistry and applications linked to biological activities of β-glucans. Nat. Prod. Rep. 2011, 28, 457–466. [Google Scholar] [CrossRef]
- Koizumi, N.; Sakagami, H.; Utsumi, A.; Fujinaga, S.; Takeda, M.; Asano, K.; Sugawara, I.; Ichikawa, S.; Kondo, H.; Mori, S.; et al. Anti-HIV (human immunodeficiency virus) activity of sulfated paramylon. Antiviral Res. 1993, 21, 1–14. [Google Scholar] [CrossRef]
- Šantek, B.; Felski, M.; Friehs, K.; Lotz, M.; Flaschel, E. Production of paramylon, a β-1,3-glucan, by heterotrophic cultivation of Euglena gracilis on potato liquor. Eng. Life Sci. 2010, 10, 165–170. [Google Scholar] [CrossRef]
- Šantek, B.; Friehs, K.; Lotz, M.; Flaschel, E. Production of paramylon, a β-1,3-glucan, by heterotrophic growth of Euglena gracilis on potato liquor in fed-batch and repeated-batch mode of cultivation. Eng. Life Sci. 2012, 12, 89–94. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Parra-Riofrío, G.; García-Márquez, J.; Casas-Arrojo, V.; Uribe-Tapia, E.; Abdala-Díaz, R.T. Antioxidant and cytotoxic effects on tumor cells of exopolysaccharides from Tetraselmis suecica (Kylin) butcher grown under autotrophic and heterotrophic conditions. Mar. Drugs 2020, 18, 534. [Google Scholar] [CrossRef] [PubMed]
- Casas-Arrojo, V.; Decara, J.; de los Ángeles Arrojo-Agudo, M.; Pérez-Manríquez, C.; Abdala-Díaz, R.T. Immunomodulatory, antioxidant activity and cytotoxic effect of sulfated polysaccharides from Porphyridium cruentum (S.F.Gray) Nägeli. Biomolecules 2021, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Metsoviti, M.N.; Papapolymerou, G.; Karapanagiotidis, I.T.; Katsoulas, N. Comparison of growth rate and nutrient content of five microalgae species cultivated in greenhouses. Plants 2019, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Tzovenis, I.; Fountoulaki, E.; Dolapsakis, N.; Kotzamanis, I.; Nengas, I.; Bitis, I.; Cladas, Y.; Economou-Amilli, A. Screening for marine nanoplanktic microalgae from Greek coastal lagoons (Ionian Sea) for use in mariculture. J. Appl. Phycol. 2009, 21, 457–469. [Google Scholar] [CrossRef]
- Órpez, R.; Martínez, M.E.; Hodaifa, G.; El Yousfi, F.; Jbari, N.; Sánchez, S. Growth of the microalga Botryococcus braunii in secondarily treated sewage. Desalination 2009, 246, 625–630. [Google Scholar] [CrossRef]
- He, J.; Xu, Y.; Chen, H.; Sun, P. Extraction, structural characterization, and potential antioxidant activity of the polysaccharides from four seaweeds. Int. J. Mol. Sci. 2016, 17, 1988. [Google Scholar] [CrossRef]
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann. Transl. Med. 2014, 2, 17. [Google Scholar] [CrossRef]
- Bhattad, T.; Koradiya, A.; Prakash, G. Prebiotic activity of paramylon isolated from heterotrophically grown Euglena gracilis. Heliyon 2021, 7, e07884. [Google Scholar] [CrossRef]
- Shanura Fernando, I.P.; Asanka Sanjeewa, K.K.; Samarakoon, K.W.; Lee, W.W.; Kim, H.S.; Kim, E.A.; Gunasekara, U.K.; Abeytunga, D.T.U.; Nanayakkara, C.; De Silva, E.D.; et al. FTIR characterization and antioxidant activity of water soluble crude polysaccharides of Sri Lankan marine algae. Algae 2017, 32, 75–86. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; Hussein, M.H.; Shaaban-Dessuuki, S.A.; Dalal, S.R. Production, extraction and characterization of Chlorella vulgaris soluble polysaccharides and their applications in AgNPs biosynthesis and biostimulation of plant growth. Sci. Rep. 2020, 10, 3011. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, F.; Wang, X.; Liu, X.; Hou, Y.; Zhang, Q. Extraction of the polysaccharides from five algae and their potential antioxidant activity in vitro. Carbohydr. Polym. 2010, 82, 118–121. [Google Scholar] [CrossRef]
- Pereira, L.; Sousa, A.; Coelho, H.; Amado, A.M.; Ribeiro-Claro, P.J.A. Use of FTIR, FT-Raman and 13C-NMR spectroscopy for identification of some seaweed phycocolloids. Biomol. Eng. 2003, 20, 223–228. [Google Scholar] [CrossRef]
- Shi, F.; Yan, X.; Cheong, K.L.; Liu, Y. Extraction, purification, and characterization of polysaccharides from marine algae Gracilaria lemaneiformis with anti-tumor activity. Process Biochem. 2018, 73, 197–203. [Google Scholar] [CrossRef]
- Meng, Y.; Yao, C.; Xue, S.; Yang, H. Application of fourier transform infrared (FT-IR) spectroscopy in determination of microalgal compositions. Bioresour. Technol. 2014, 151, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Barras, D.R.; Stone, B.A. Chemical composition of pellicle of Euglena gracilis var Bacillaris. Biochem. J. 1965, 97, 14–15. [Google Scholar]
- Bouck, G.B.; Rogalski, A.; Valaitis, A. Surface organization and composition of Euglena. 2. Flagellar mastigonemes. J. Cell Biol. 1978, 77, 805–826. [Google Scholar] [CrossRef]
- Phillips, C.; Jensen, G.; Showman, L.; Tonda, R.; Horst, G.; Levine, R. Particulate and solubilized β-glucan and non-βglucan fractions of Euglena gracilis induce pro- and anti-inflammatory innate immune cell responses and exhibit antioxidant properties. J. Inflamm. Res. 2019, 12, 49–64. [Google Scholar] [CrossRef]
- Assunção, M.F.G.; Amaral, R.; Martins, C.B.; Ferreira, J.D.; Ressurreição, S.; Santos, S.D.; Varejão, J.M.T.B.; Santos, L.M.A. Screening microalgae as potential sources of antioxidants. J. Appl. Phycol. 2017, 29, 865–877. [Google Scholar] [CrossRef]
- Li, H.B.; Cheng, K.W.; Wong, C.C.; Fan, K.W.; Chen, F.; Yue, J. Evaluation of antioxidant capacity and total phenolic content of different fractions of selected microalgae. Food Chem. 2007, 102, 771–776. [Google Scholar] [CrossRef]
- Guedes, A.C.; Amaro, H.M.; Gião, M.S.; Malcata, F.X. Optimization of ABTS radical cation assay specifically for determination of antioxidant capacity of intracellular extracts of microalgae and cyanobacteria. Food Chem. 2013, 138, 638–643. [Google Scholar] [CrossRef]
- Guedes, A.C.; Amaro, H.M.; Malcata, F.X. Microalgae as sources of high added-value compounds—A brief review of recent work. Biotechnol. Prog. 2011, 27, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Graziani, G.; Schiavo, S.; Nicolai, M.A.; Buono, S.; Fogliano, V.; Pinto, G.; Pollio, A. Microalgae as human food: Chemical and nutritional characteristics of the thermo-acidophilic microalga Galdieria sulphuraria. Food Funct. 2013, 4, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Emad, A.S.; Sanaa, M.M.S.; Vikramjit, S. Salt stress enhancement of antioxidant and antiviral efficiency of Spirulina platensis. J. Med. Plants Res. 2010, 4, 2622–2632. [Google Scholar] [CrossRef]
- Pellegrini, N.; Serafini, M.; Colombi, B.; Del Rio, D.; Salvatore, S.; Bianchi, M.; Brighenti, F. Total antioxidant capacity of plant foods, beverages and oils consumed in Italy assessed by three different in vitro assays. J. Nutr. 2003, 133, 2812–2819. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, E.A.; Shanab, S.M.M. Comparison of DPPH and ABTS assays for determining antioxidant potential of water and methanol extracts of Spirulina platensis. Indian J. Mar. Sci. 2013, 42, 556–564. [Google Scholar]
- Bayona, K.; Navarro, G.S.M.; Lara, A.D.; Colorado, J.; Atehortúa, L.; Martínez, A. Activity of sulfated polysaccharides from microalgae Porphyridium cruentum over degenerative mechanisms of the skin. Int. J. Sci. Adv. Technol. 2012, 2, 85–92. [Google Scholar]
- Abd El Baky, H.; Hanaa El Baz, K.F.; EL-Latife, S.A. Induction of sulfated polysaccharides in Spirulina platensis as response to nitrogen concentration and its biological evaluation. J. Aquac. Res. Dev. 2016, 5, 206. [Google Scholar] [CrossRef]
- Tsiapali, E.; Whaley, S.; Kalbfleisch, J.; Ensley, H.E.; Browder, I.W.; Williams, D.L. Glucans exhibit weak antioxidant activity, but stimulate macrophage free radical activity. Free Radic. Biol. Med. 2001, 30, 393–402. [Google Scholar] [CrossRef]
- Tannin-Spitz, T.; Bergman, M.; Van-Moppes, D.; Grossman, S.; Arad, S. Antioxidant activity of the polysaccharide of the red microalga Porphyridium sp. J. Appl. Phycol. 2005, 17, 215–222. [Google Scholar] [CrossRef]
- Leung, P.H.; Zhao, S.; Ho, K.P.; Wu, J.Y. Chemical properties and antioxidant activity of exopolysaccharides from mycelial culture of Cordyceps sinensis fungus Cs-HK1. Food Chem. 2009, 114, 1251–1256. [Google Scholar] [CrossRef]
- Yan, J.; Wang, W.; Ma, H.; Wu, J. Sulfation and enhanced antioxidant capacity of an exopolysaccharide produced by the medicinal fungus Cordyceps sinensis. Molecules 2013, 18, 167–177. [Google Scholar] [CrossRef] [PubMed]
- De Jesus Raposo, F.M.; Bernardo De Morais, A.M.; Santos Costa de Morais, R.M. Marine polysaccharides from algae with potential biomedical applications. Mar. Drugs 2015, 13, 2967–3028. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, L.; Wang, H.; Liu, S.; Yu, H.; Wang, X.; Li, R.; Liu, T.; Li, P. Partial characterization, the immune modulation and anticancer activities of sulfated polysaccharides from filamentous microalgae Tribonema sp. Molecules 2019, 24, 322. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wan, H.; Wang, R.; Hao, D. Sulfated polysaccharides from Phaeodactylum tricornutum: Isolation, structural characteristics, and inhibiting HepG2 growth activity in vitro. PeerJ 2019, 7, e6409. [Google Scholar] [CrossRef]
- Umemura, K.; Yanase, K.; Suzuki, M.; Okutani, K.; Yamori, T.; Andoh, T. Inhibition of DNA topoisomerases I and II, and growth inhibition of human cancer cell lines by a marine microalgal polysaccharide. Biochem. Pharmacol. 2003, 66, 481–487. [Google Scholar] [CrossRef]
- Castro, R.; Piazzon, M.C.; Zarra, I.; Leiro, J.; Noya, M.; Lamas, J. Stimulation of turbot phagocytes by Ulva rigida C. Agardh polysaccharides. Aquaculture 2006, 254, 9–20. [Google Scholar] [CrossRef]
- Schepetkin, I.A.; Quinn, M.T. Botanical polysaccharides: Macrophage immunomodulation and therapeutic potential. Int. Immunopharmacol. 2006, 6, 317–333. [Google Scholar] [CrossRef]
- Pereira, B.M.R.; da Silva, B.P.; Pereira, N.A.; Parente, J.P. Anti-inflammatory and immunologically active polysaccharides of Periandra mediterranea. Phytochemistry 2000, 54, 409–413. [Google Scholar] [CrossRef]
- Ann-Chang Cheng, C.-W.T. The immunostimulatory effects of sodium alginate and iota-carrageenan on orange-spotted grouper Epinephelus coicoides and its resistance against Vibrio alginolyticus. Fish Shellfish Immunol. 2007, 22, 197–205. [Google Scholar] [CrossRef]
- Abdala Díaz, R.T.; Chabrillón, M.; Cabello Pasini, A.; López Soler, B.; Figueroa, F.L. Effect of Porphyridium cruentum polysaccharides on the activity of murine macrophage cell line RAW 264.7. Ciencias Mar. 2010, 36, 345–353. [Google Scholar] [CrossRef]
- Abdala Díaz, R.T.; Casas Arrojo, V.; Arrojo Agudo, M.A.; Cárdenas, C.; Dobretsov, S.; Figueroa, F.L. Immunomodulatory and antioxidant activities of sulfated polysaccharides from Laminaria ochroleuca, Porphyra umbilicalis and Gelidium corneum. Mar. Biotechnol. 2019, 21, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.Q. Preparation of Polysaccharides from Porphyridium cruentum and Their Biological Activities. Ph.D. Thesis, Dalian University of Technology, Dalian, China, 2010. [Google Scholar]
- Ketteler, M.; Bongartz, P.; Westenfeld, R.; Wildberger, J.E.; Mahnken, A.H.; Böhm, R.; Metzger, T.; Wanner, C.; Jahnen-dechent, W.; Floege, J. Association of low fetuin-A (AHSG) concentrations in serum with cardiovascular mortality in patients on dialysis: A cross-sectional study. Lancet 2003, 361, 827–833. [Google Scholar] [CrossRef]
- Webb, R. Smooth muscle contraction and relaxation. Adv. Physiol. Educ. 2003, 27, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, M.; Kimura, M.Y.; Ito, T.; Hayashizaki, K.; Endo, Y.; Wang, Y.; Yagi, R.; Nakagawa, T.; Kato, N.; Matsubara, H.; et al. Myosin light chain 9/12 regulates the pathogenesis of inflammatory bowel disease. Front. Immunol. 2021, 11, 594297. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Tran, G.B.; Nguyen, C.T. Anti-oxidative effects of superoxide dismutase 3 on inflammatory diseases. J. Mol. Med. 2020, 98, 59–69. [Google Scholar] [CrossRef]
- Sharma, S.; Rais, A.; Sandhu, R.; Nel, W.; Ebadi, M. Clinical significance of metallothioneins in cell therapy and nanomedicine. Int. J. Nanomed. 2013, 8, 1477–1488. [Google Scholar] [CrossRef]
- Maret, W. Redox biochemistry of mammalian metallothioneins. J. Biol. Inorg. Chem. 2011, 16, 1079–1086. [Google Scholar] [CrossRef]
- Miura, T.; Muraoka, S.; Ogiso, T. Antioxidant activity of metallothionein compared with reduced glutathione. Life Sci. 1997, 60, 301–309. [Google Scholar] [CrossRef]
- Vignesh, K.S.; Deepe, G.S. Metallothioneins: Emerging modulators in immunity and infection. Int. J. Mol. Sci. 2017, 18, 2197. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Itoh, N.; Kawasaki, A.; Matsuyama, A.; Matsuda, K.; Nakanishi, T.; Tanaka, K. Metallothionein modulates lipopolysaccharide-stimulated tumour necrosis factor expression in mouse peritoneal macrophages. Biochem. J. 2002, 361, 363–369. [Google Scholar] [CrossRef]
- Hall, A.K. Thymosin beta-10 accelerates apoptosis. Cell. Mol. Biol. Res. 1995, 41, 167–180. [Google Scholar] [PubMed]
- Lourenço, S.O.; Barbarino, E.; Lavín, P.L.; Lanfer Marquez, U.M.; Aidar, E. Distribution of intracellular nitrogen in marine microalgae: Calculation of new nitrogen-to-protein conversion factors. Eur. J. Phycol. 2004, 39, 17–32. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Biochem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Kochert, A.G. Carbohydrate Determination by the Phenol–Sulfuric Acid Method. In Handbook of Phycological Methods, Phycological and Biochemical Methods; Cambridge University Press: Cambridge, UK, 1978. [Google Scholar]
- Folch, J.; Less, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Parages, M.L.; Rico, R.M.; Abdala-Díaz, R.T.; Chabrillón, M.; Sotiroudis, T.G.; Jiménez, C. Acidic polysaccharides of Arthrospira (Spirulina) platensis induce the synthesis of TNF-α in RAW macrophages. J. Appl. Phycol. 2012, 24, 1537–1546. [Google Scholar] [CrossRef]
- Abdala Díaz, R.T.; Chabrillón, M.; Cabello-Pasini, A.; Gómez-Pinchetti, J.L.; Figueroa, F.L. Characterization of polysaccharides from Hypnea spinella (Gigartinales) and Halopithys incurva (Ceramiales) and their effect on RAW 264.7 macrophage activity. J. Appl. Phycol. 2010, 23, 523–528. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Vijayabaskar, P.; Vaseela, N. In vitro antioxidant properties of sulfated polysaccharide from brown marine algae Sargassum tenerrimum. Asian Pacific J. Trop. Dis. 2012, 2, S890–S896. [Google Scholar] [CrossRef]
- Brand-Willians, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- JK, E.; AL, M.; JR, Y. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]










| Biomass (%) | Extracted Polysaccharides (%) | |
|---|---|---|
| Carbon | 44.59 ± 0.97 | 1.24 ± 0.82 |
| Hydrogen | 5.57 ± 0.41 | 0.20 ± 0.09 |
| Nitrogen | 10.54 ± 1.02 | 0.24 ± 0.07 |
| Sulphur | 0.03 ± 0.003 | 0.16 ± 0.06 |
| Proteins | 52.59± 0.69% |
| Carbohydrates | 7.98 ± 1.23% |
| Lipids | 11.30 ± 0.72% |
| Inorganic compounds | 11.64 ± 1.38 % |
| Moisture | 16.49 ± 1.98% |
| Header | Monosaccharide | Retention Time (min) | Peak Area | % Mass |
|---|---|---|---|---|
| 1 | Ribose (Rib) | 19.19 | 122,217,046 | 19.58 |
| 2 | Fucose (Fuc) | 20.28 | 29,872,029 | 4.79 |
| 3 | Mannose (Mann) | 26.94 | 113,488,499 | 18.18 |
| 4 | Galactose (Gal) | 28.20 | 377,111,564 | 6.04 |
| 5 | Glucose (Glc) | 29.73 | 21,748,410 | 34.83 |
| UniProt Accession | Gene Symbol | Description | Sum PEP Score * | Abundance Ratio: Treatment/Control | Abundance Ratio p-Value |
|---|---|---|---|---|---|
| O14950 | MYL12B; MYL12A | Myosin regulatory light chain 12A/B | 81.34 | 1.34 | 2.96 × 10−2 |
| P04179 | SOD2 | Superoxide dismutase [Mn], mitochondrial | 30.42 | 1.44 | 2.48 × 10−6 |
| P02795 | MT2A | Metallothionein-2 | 28.35 | 1.23 | 1.90 × 10−44 |
| P02765 | AHSG | Alpha-2-HS-glycoprotein | 16.59 | 1.62 | 4.83 × 10−22 |
| P63313 | TMSB10 | Thymosin beta-10 | 13.29 | 1.33 | 3.32 × 10−22 |
| Node 1 | Node 2 | Score |
|---|---|---|
| AHSG | IL6 | 0.518 |
| AHSG | TNF | 0.474 |
| IL6 | AHSG | 0.518 |
| IL6 | MT2A | 0.344 |
| IL6 | SOD2 | 0.594 |
| IL6 | TNF | 0.994 |
| MT2A | IL6 | 0.344 |
| MT2A | SOD2 | 0.336 |
| MT2A | TMSB10 | 0.173 |
| MT2A | TNF | 0.281 |
| MYL12B | TMSB10 | 0.201 |
| SOD2 | IL6 | 0.594 |
| SOD2 | MT2A | 0.336 |
| SOD2 | TNF | 0.619 |
| TMSB10 | MT2A | 0.173 |
| TMSB10 | MYL12B | 0.201 |
| TMSB10 | TNF | 0.181 |
| TNF | AHSG | 0.474 |
| TNF | IL6 | 0.994 |
| TNF | MT2A | 0.281 |
| TNF | SOD2 | 0.619 |
| TNF | TMSB10 | 0.181 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casas-Arrojo, V.; Arrojo Agudo, M.d.l.Á.; Cárdenas García, C.; Carrillo, P.; Pérez Manríquez, C.; Martínez-Manzanares, E.; Abdala Díaz, R.T. Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs. Pharmaceuticals 2022, 15, 1379. https://doi.org/10.3390/ph15111379
Casas-Arrojo V, Arrojo Agudo MdlÁ, Cárdenas García C, Carrillo P, Pérez Manríquez C, Martínez-Manzanares E, Abdala Díaz RT. Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs. Pharmaceuticals. 2022; 15(11):1379. https://doi.org/10.3390/ph15111379
Chicago/Turabian StyleCasas-Arrojo, Virginia, María de los Ángeles Arrojo Agudo, Casimiro Cárdenas García, Paloma Carrillo, Claudia Pérez Manríquez, Eduardo Martínez-Manzanares, and Roberto T. Abdala Díaz. 2022. "Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs" Pharmaceuticals 15, no. 11: 1379. https://doi.org/10.3390/ph15111379
APA StyleCasas-Arrojo, V., Arrojo Agudo, M. d. l. Á., Cárdenas García, C., Carrillo, P., Pérez Manríquez, C., Martínez-Manzanares, E., & Abdala Díaz, R. T. (2022). Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs. Pharmaceuticals, 15(11), 1379. https://doi.org/10.3390/ph15111379







