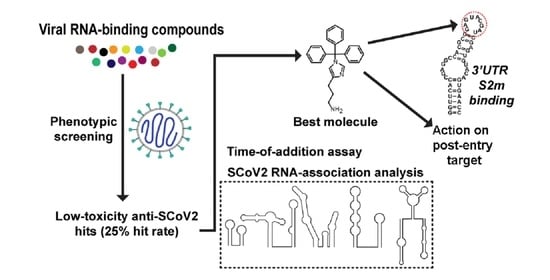
SARS-CoV-2 Inhibitors Identified by Phenotypic Analysis of a Collection of Viral RNA-Binding Molecules
Abstract
:1. Introduction
2. Results
2.1. Selection of Compounds for the Phenotypic Screen
2.2. Anti-SCoV2 Activities and Selectivity Indexes
2.3. Time-of-Addition Assays
2.4. SCoV2 RNA-Binding Experiments
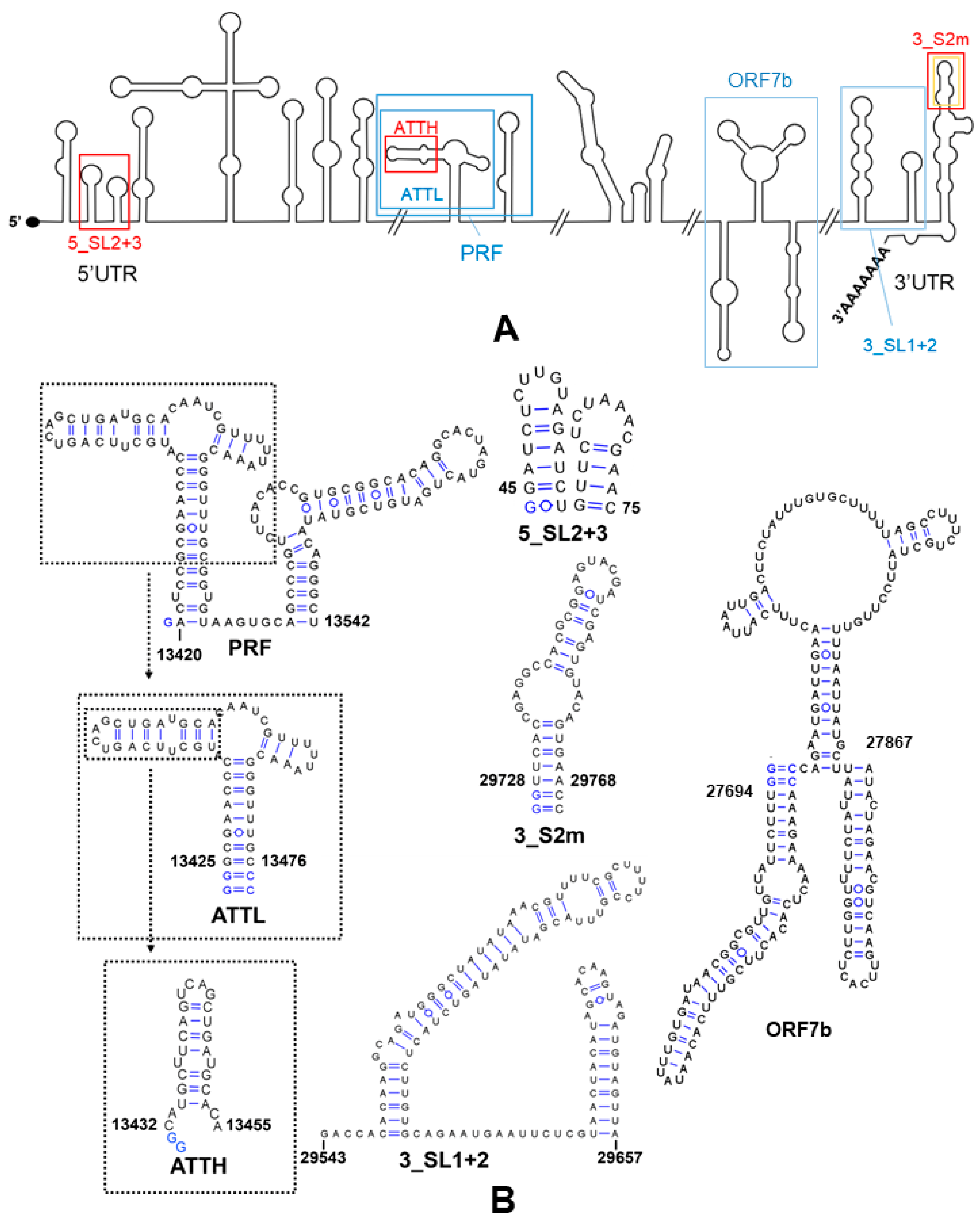
2.4.1. Selection of RNA Sequences
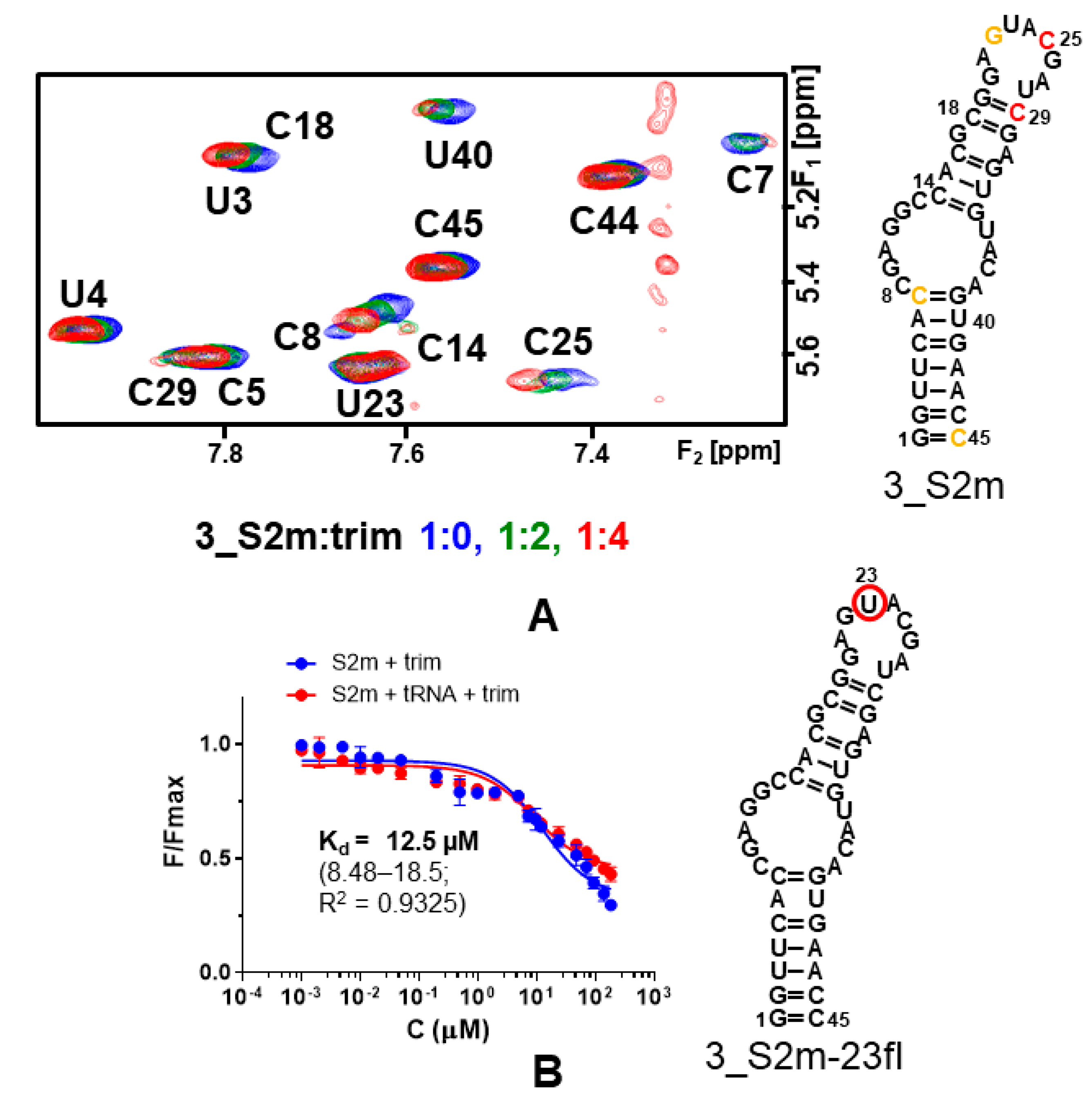
2.4.2. Ligand-Based NMR Experiments
2.4.3. RNA-Based NMR Experiments
2.4.4. Fluorescence Binding Assays
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Evaluation of Anti-SCoV2 Activity and Cellular Toxicity
4.3. Time-of-Addition Assays
4.4. Preparation of SCoV2 RNA Samples for NMR Spectroscopy and Fluorescence Experiments
4.5. NMR Spectroscopy
4.5.1. Ligand-Based Experiments
4.5.2. RNA-Based Experiments
4.6. Fluorescence Binding Assays
4.7. Molecular Modeling
5. Patents
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tao, K.; Tzou, P.L.; Nouhin, J.; Bonilla, H.; Jagannathan, P.; Shafer, R.W. SARS-CoV-2 Antiviral Therapy. Clin. Microbiol. Rev. 2021, 34, e0010921. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Hajdin, C.E.; Weeks, K.M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018, 17, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Wacker, A.; Weigand, J.E.; Akabayov, S.R.; Altincekic, N.; Bains, J.K.; Banijamali, E.; Binas, O.; Castillo-Martinez, J.; Cetiner, E.; Ceylan, B.; et al. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. 2020, 48, 12415–12435. [Google Scholar] [CrossRef] [PubMed]
- Prado, S.; Beltrán, M.; Coiras, M.; Bedoya, L.M.; Alcamí, J.; Gallego, J. Bioavailable inhibitors of HIV-1 RNA biogenesis identified through a Rev-based screen. Biochem. Pharmacol. 2016, 107, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Prado, S.; Beltrán, M.; Moreno, A.; Bedoya, L.M.; Alcamí, J.; Gallego, J. A small-molecule inhibitor of HIV-1 Rev function detected by a diversity screen based on RRE-Rev interference. Biochem. Pharmacol. 2018, 156, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Simba-Lahuasi, A.; Bedoya, L.M.; Alcami, J.; Gallego, J. Novel HIV-1 RNA biogenesis inhibitors identified by a virtual pharmacophore-based search. Unpublished Results. 2022. [Google Scholar]
- Gonzalez-Bulnes, L.; Ibanez, I.; Bedoya, L.M.; Beltran, M.; Catalan, S.; Alcami, J.; Fustero, S.; Gallego, J. Structure-Based Design of an RNA-Binding p-Terphenylene Scaffold that Inhibits HIV-1 Rev Protein Function. Angew. Chem.-Int. Ed. 2013, 52, 13405–13409. [Google Scholar] [CrossRef]
- Medina-Trillo, C.; Sedgwick, D.M.; Herrera, L.; Beltrán, M.; Moreno, A.; Barrio, P.; Bedoya, L.M.; Alcamí, A.; Fustero, S.; Gallego, J. Nucleic acid recognition and antiviral activity of 1,4-substituted terphenyl compounds mimicking all faces of the HIV-1 Rev protein positively-charged alpha-helix. Sci. Rep. 2020, 10, 7190. [Google Scholar] [CrossRef]
- Martín-Villamil, M.; Sanmartín, I.; Moreno, A.; Gallego, J. Pharmacophore-based discovery of viral RNA conformational modulators. Pharmaceuticals 2022, 15, 748. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Miyaji, A.; Jefferson, E.A.; Sannes-Lowery, K.A.; Osgood, S.A.; Propp, S.S.; Ranken, R.; Massire, C.; Sampath, R.; Ecker, D.J.; et al. SAR by MS: Discovery of a new class of RNA-binding small molecules for the hepatitis C virus: Internal ribosome entry site IIA subdomain. J. Med. Chem. 2005, 48, 7099–7102. [Google Scholar] [CrossRef] [PubMed]
- Madhugiri, R.; Karl, N.; Petersen, D.; Lamkiewicz, K.; Fricke, M.; Wend, U.; Scheuer, R.; Marz, M.; Ziebuhr, J. Structural and functional conservation of cis-acting RNA elements in coronavirus 5′-terminal genome regions. Virology 2018, 517, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.A.; Woodside, M.T.; Dinman, J.D. Programmed -1 Ribosomal Frameshifting in coronaviruses: A therapeutic target. Virology 2021, 554, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; DeFalco, L.; Anderson, D.E.; Zhang, Y.; Aw, J.G.A.; Lim, S.Y.; Lim, X.N.; Tan, K.Y.; Zhang, T.; Chawla, T.; et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 2021, 12, 5113. [Google Scholar] [CrossRef]
- Goebel, S.J.; Hsue, B.; Dombrowski, T.F.; Masters, P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004, 78, 669–682. [Google Scholar] [CrossRef] [Green Version]
- Imperatore, J.A.; Cunningham, C.L.; Pellegrene, K.A.; Brinson, R.G.; Marino, J.P.; Evanseck, J.D.; Mihailescu, M.R. Highly conserved s2m element of SARS-CoV-2 dimerizes via a kissing complex and interacts with host miRNA-1307-3p. Nucleic Acids Res. 2022, 50, 1017–1032. [Google Scholar] [CrossRef]
- Manfredonia, I.; Nithin, C.; Ponce-Salvatierra, A.; Ghosh, P.; Wirecki, T.K.; Marinus, T.; Ogando, N.S.; Snijder, E.J.; van Hemert, M.J.; Bujnicki, J.M.; et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020, 48, 12436–12452. [Google Scholar] [CrossRef]
- Lan, T.C.T.; Allan, M.F.; Malsick, L.E.; Woo, J.Z.; Zhu, C.; Zhang, F.; Khandwala, S.; Nyeo, S.S.Y.; Sun, Y.; Guo, J.U.; et al. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat. Commun. 2022, 13, 1128. [Google Scholar] [CrossRef]
- Calabrese, D.R.; Connelly, C.M.; Schneekloth, J.S. Ligand-observed NMR techniques to probe RNA-small molecule interactions. Methods Enzymol. 2019, 623, 131–149. [Google Scholar] [CrossRef]
- Sreeramulu, S.; Richter, C.; Berg, H.; Wirtz Martin, M.A.; Ceylan, B.; Matzel, T.; Adam, J.; Altincekic, N.; Azzaoui, K.; Bains, J.K.; et al. Exploring the Druggability of Conserved RNA Regulatory Elements in the SARS-CoV-2 Genome. Angew. Chem.-Int. Ed. Engl. 2021, 60, 19191–19200. [Google Scholar] [CrossRef]
- Kumari, M.; Lu, R.M.; Li, M.C.; Huang, J.L.; Hsu, F.F.; Ko, S.H.; Ke, F.Y.; Su, S.C.; Liang, K.H.; Yuan, J.P.; et al. A critical overview of current progress for COVID-19: Development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci. 2022, 29, 68. [Google Scholar] [CrossRef]
- Ng, T.I.; Correia, I.; Seagal, J.; DeGoey, D.A.; Schrimpf, M.R.; Hardee, D.J.; Noey, E.L.; Kati, W.M. Antiviral Drug Discovery for the Treatment of COVID-19 Infections. Viruses 2022, 14, 961. [Google Scholar] [CrossRef] [PubMed]
- Puhl, A.C.; Lane, T.R.; Urbina, F.; Ekins, S. The Need for Speed and Efficiency: A Brief Review of Small Molecule Antivirals for COVID-19. Front. Drug Discov. 2022, 2, 837587. [Google Scholar] [CrossRef]
- Nepali, K.; Sharma, R.; Sharma, S.; Thakur, A.; Liou, J.P. Beyond the vaccines: A glance at the small molecule and peptide-based anti-COVID19 arsenal. J. Biomed. Sci. 2022, 29, 65. [Google Scholar] [CrossRef]
- Haniff, H.S.; Tong, Y.; Liu, X.; Chen, J.L.; Suresh, B.M.; Andrews, R.J.; Peterson, J.M.; O’Leary, C.A.; Benhamou, R.I.; Moss, W.N.; et al. Targeting the SARS-CoV-2 RNA Genome with Small Molecule Binders and Ribonuclease Targeting Chimera (RIBOTAC) Degraders. ACS Cent. Sci. 2020, 6, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Zafferani, M.; Haddad, C.; Luo, L.; Davila-Calderon, J.; Yuan-Chiu, L.; Shema Mugisha, C.; Monaghan, A.G.; Kennedy, A.A.; Yesselman, J.D.; Gifford, R.R.; et al. Amilorides inhibit SARS-CoV-2 replication in vitro by targeting RNA structures. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Vora, S.M.; Fontana, P.; Mao, T.; Leger, V.; Zhang, Y.; Fu, T.M.; Lieberman, J.; Gehrke, L.; Shi, M.; Wang, L.; et al. Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion. Proc. Natl. Acad. Sci. USA 2022, 119. [Google Scholar] [CrossRef]
- Hegde, S.; Tang, Z.; Zhao, J.; Wang, J. Inhibition of SARS-CoV-2 by Targeting Conserved Viral RNA Structures and Sequences. Front. Chem. 2021, 9, 802766. [Google Scholar] [CrossRef]
- Sun, Y.; Abriola, L.; Niederer, R.O.; Pedersen, S.F.; Alfajaro, M.M.; Silva Monteiro, V.; Wilen, C.B.; Ho, Y.C.; Gilbert, W.V.; Surovtseva, Y.V.; et al. Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Xiong, H.L.; Cao, J.L.; Shen, C.G.; Ma, J.; Qiao, X.Y.; Shi, T.S.; Ge, S.X.; Ye, H.M.; Zhang, J.; Yuan, Q.; et al. Several FDA-Approved Drugs Effectively Inhibit SARS-CoV-2 Infection in vitro. Front. Pharmacol. 2020, 11, 609592. [Google Scholar] [CrossRef]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 2020, 64, e00819-20. [Google Scholar] [CrossRef] [PubMed]
- Tummino, T.A.; Rezelj, V.V.; Fischer, B.; Fischer, A.; O'Meara, M.J.; Monel, B.; Vallet, T.; White, K.M.; Zhang, Z.; Alon, A.; et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 2021, 373, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Rangan, R.; Watkins, A.M.; Chacon, J.; Kretsch, R.; Kladwang, W.; Zheludev, I.N.; Townley, J.; Rynge, M.; Thain, G.; Das, R. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res. 2021, 49, 3092–3108. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; White, K.M.; Miorin, L.; Strohmeier, S.; McMahon, M.; Meade, P.; Liu, W.C.; Albrecht, R.A.; Simon, V.; Martinez-Sobrido, L.; et al. An In Vitro Microneutralization Assay for SARS-CoV-2 Serology and Drug Screening. Curr. Protoc. Microbiol. 2020, 58, e108. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [Green Version]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef]
- Molle, I.; Thomann, A.; Buckley, D.L.; So, E.C.; Lang, S.; Crews, C.M.; Ciulli, A. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein:hypoxia inducible factor 1sigma protein-protein interface. Chem. Biol. 2012, 19, 1300–1312. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.W.; Li, L.; Giedroc, D.P. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 2011, 585, 1049–1053. [Google Scholar] [CrossRef] [Green Version]
- Bradrick, T.D.; Marino, J.P. Ligand-induced changes in 2-aminopurine fluorescence as a probe for small molecule binding to HIV-1 TAR RNA. RNA 2004, 10, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simba-Lahuasi, A.; Cantero-Camacho, Á.; Rosales, R.; McGovern, B.L.; Rodríguez, M.L.; Marchán, V.; White, K.M.; García-Sastre, A.; Gallego, J. SARS-CoV-2 Inhibitors Identified by Phenotypic Analysis of a Collection of Viral RNA-Binding Molecules. Pharmaceuticals 2022, 15, 1448. https://doi.org/10.3390/ph15121448
Simba-Lahuasi A, Cantero-Camacho Á, Rosales R, McGovern BL, Rodríguez ML, Marchán V, White KM, García-Sastre A, Gallego J. SARS-CoV-2 Inhibitors Identified by Phenotypic Analysis of a Collection of Viral RNA-Binding Molecules. Pharmaceuticals. 2022; 15(12):1448. https://doi.org/10.3390/ph15121448
Chicago/Turabian StyleSimba-Lahuasi, Alvaro, Ángel Cantero-Camacho, Romel Rosales, Briana Lynn McGovern, M. Luis Rodríguez, Vicente Marchán, Kris M. White, Adolfo García-Sastre, and José Gallego. 2022. "SARS-CoV-2 Inhibitors Identified by Phenotypic Analysis of a Collection of Viral RNA-Binding Molecules" Pharmaceuticals 15, no. 12: 1448. https://doi.org/10.3390/ph15121448
APA StyleSimba-Lahuasi, A., Cantero-Camacho, Á., Rosales, R., McGovern, B. L., Rodríguez, M. L., Marchán, V., White, K. M., García-Sastre, A., & Gallego, J. (2022). SARS-CoV-2 Inhibitors Identified by Phenotypic Analysis of a Collection of Viral RNA-Binding Molecules. Pharmaceuticals, 15(12), 1448. https://doi.org/10.3390/ph15121448