Targeting the DNA Damage Response Machinery for Lung Cancer Treatment
Abstract
:1. Introduction
2. DNA Repair Pathways for the Targeting of Lung Cancer
2.1. Homologous Recombination (HR) Pathway
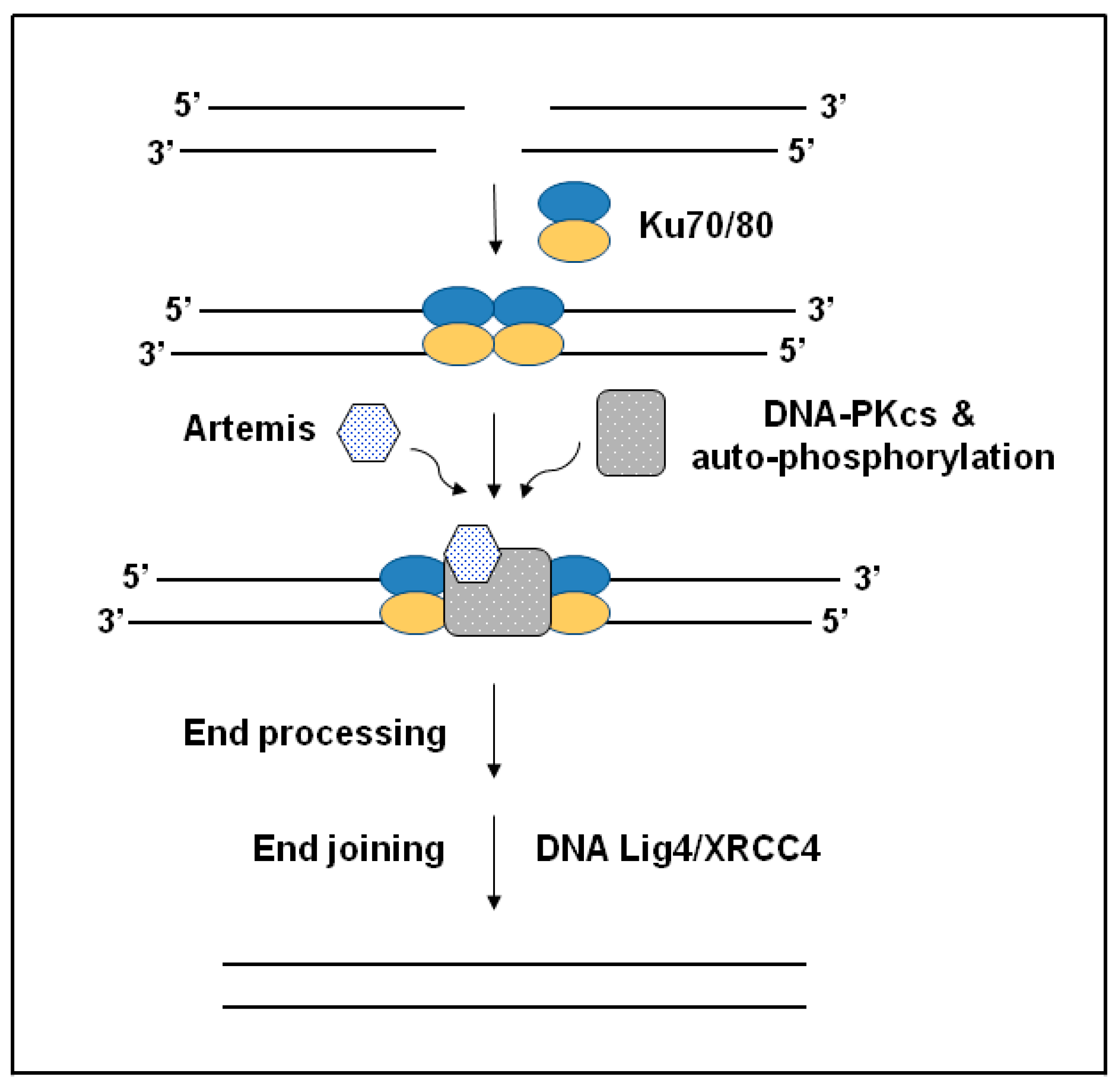
2.2. Non-Homologous End Joining (NHEJ) Pathway
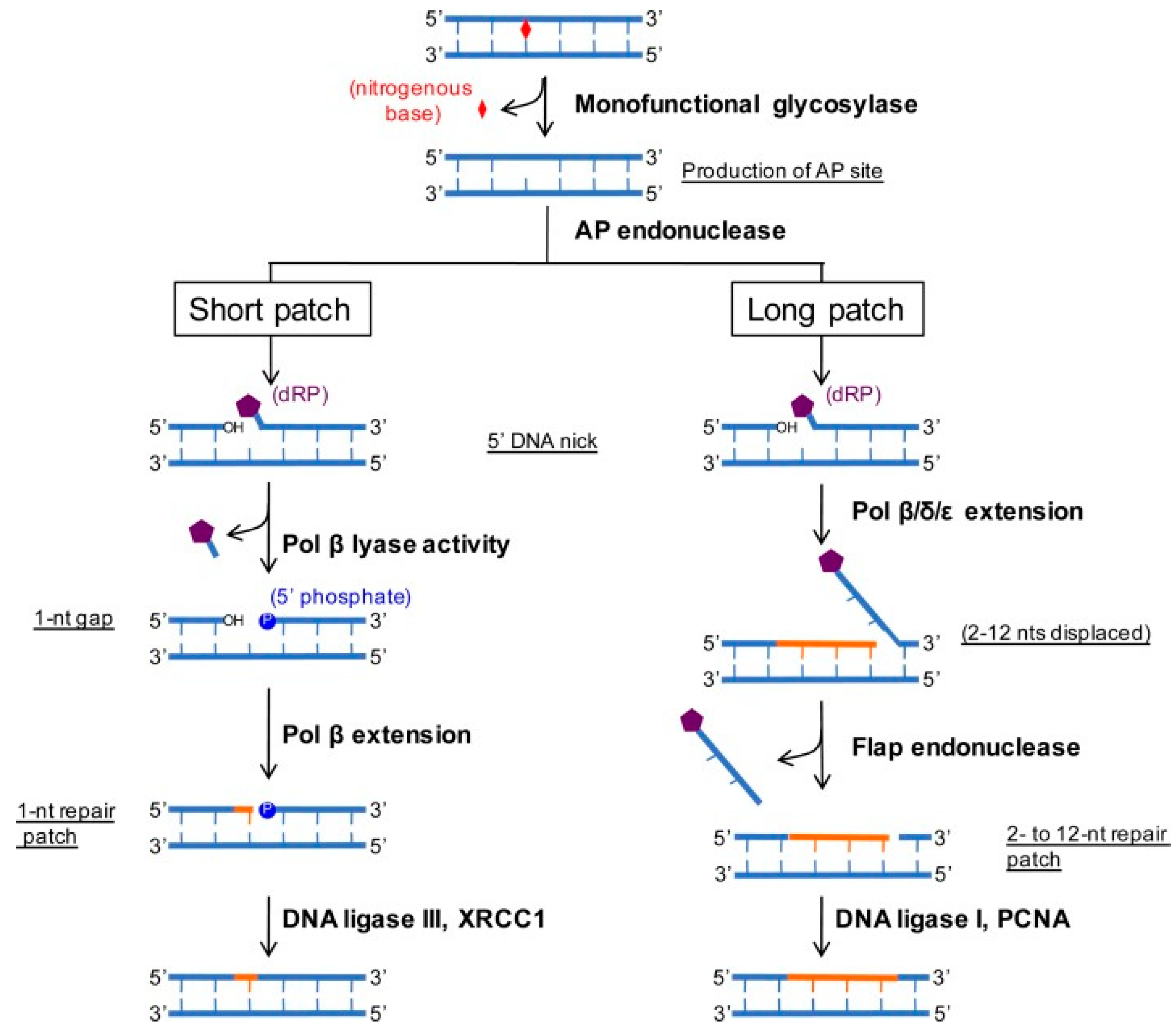
2.3. Base Excision Repair (BER) Pathway
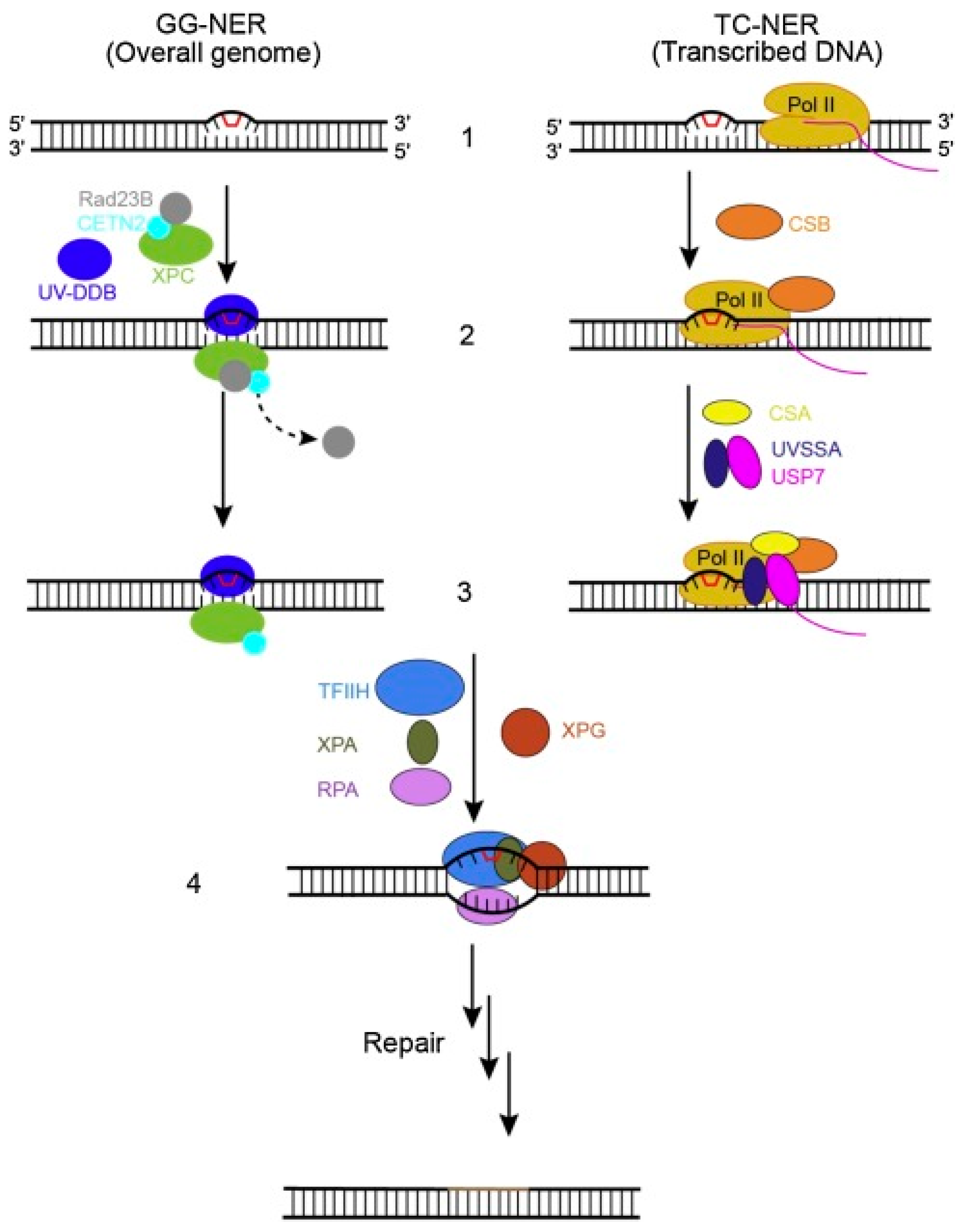
2.4. Nucleotide Excision Repair (NER) Pathway
3. DDR Inhibitors for a Targeted Treatment of Lung Cancer
3.1. ATM/ATR Inhibitors
3.2. DNA-PK Inhibitors
3.3. PARP Inhibitors
4. The Current Status and Future Perspectives
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATM | Ataxia telangiectasia mutated |
| ATR | Rad3-related protein |
| ATRIP | ATR interacting protein |
| BER | Base excision repair |
| COPD | Chronic obstructive pulmonary disease |
| DDR | DNA damage response |
| DNA-PK | DNA-dependent protein kinase |
| DNAPKcs | DNA-dependent protein kinase catalytic subunit |
| DSBs | Double-strand breaks |
| EGFR-TKIs | EGFR tyrosine kinase inhibitors |
| ERCC | Excision repair cross complementation |
| GG-NER | Global genome NER |
| HR | Homologous recombination |
| HRD | Homologous recombination deficiency |
| IR | Ionizing radiation |
| LCNEC | Large cell neuroendocrine carcinoma |
| MMR | Mismatch repair |
| NER | Nucleotide excision repair |
| NHEJ | Non-homologous end joining |
| NSCLC | Non-small cell lung cancer |
| OPPs | Organophosphate pesticides |
| PARP | Poly-ADP-ribose polymerase |
| PDX | Patient-derived xenograft |
| RFA | Radiofrequency ablation |
| ROS | Reactive oxygen species |
| RPA | Replication protein A |
| RPPA | Reverse phase protein array |
| RT | Radiotherapy |
| SCLC | Small cell lung cancer |
| SLFN11 | Schlafen 11 |
| SqCCs | Squamous cell carcinomas |
| SSBs | Single-strand breaks |
| TCGA | The cancer genome atlas |
| TC-NER | Transcription-coupled NER |
| TMZ | Temozolomide |
| UV | Ultraviolet |
| WRN | Werner syndrome protein |
| XRCC4 | X-ray repair complementing defective repair in Chinese-hamster cells 4 |
References
- Stefanou, D.T.; Kouvela, M.; Stellas, D.; Voutetakis, K.; Papadodima, O.; Syrigos, K.; Souliotis, V.L. Oxidative Stress and Deregulated DNA Damage Response Network in Lung Cancer Patients. Biomedicines 2022, 10, 1248. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [Green Version]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci 2019, 21, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edge, L. Review Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [Green Version]
- Keng, P.; Faivre-finn, C.; Blackhall, F.H.; Ruysscher, D. De Targeted Agents in Non-Small Cell Lung Cancer (NSCLC ): Clinical Developments and Rationale for the Combination with Thoracic Radiotherapy. Cancer Treat. Rev. 2012, 38, 626–640. [Google Scholar] [CrossRef]
- Reuvers, T.G.A.; Kanaar, R.; Nonnekens, J. DNA Damage-Inducing Anticancer Therapies: From Global to Precision Damage. Cancers 2020, 12, 2098. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jin, G.; Cai, F.; Chen, X.; Cao, N.; Zhang, X.; Liu, J.; Chen, F.; Wang, F. Extracellular Signal-Regulated Kinase 5 Increases Radioresistance of Lung Cancer Cells by Enhancing the DNA Damage Response. Exp. Mol. Med. 2019, 51, 1–20. [Google Scholar] [CrossRef]
- Fortney, K.; Jurisica, I. Integrative Computational Biology for Cancer Research. Hum. Genet. 2011, 130, 465–481. [Google Scholar] [CrossRef] [Green Version]
- Chang, A. Lung Cancer Chemotherapy, Chemoresistance and the Changing Treatment Landscape for NSCLC. Lung Cancer 2011, 71, 3–10. [Google Scholar] [CrossRef]
- Kara, A.; Özgür, A.; Nalbantoğlu, S.; Karadağ, A. DNA Repair Pathways and Their Roles in Drug Resistance for Lung Adenocarcinoma Double Holliday Junction. Mol. Biol. Rep. 2021, 48, 3813–3825. [Google Scholar] [CrossRef]
- Cai, D.; Choi, P.S.; Gelbard, M.; Meyerson, M. Identification and Characterization of Oncogenic SOS1 Mutations in Lung Adenocarcinoma. Mol. Cancer Res. 2019, 17, 617–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouwman, P.; Jonkers, J. Genomic Instability in Cancer: The Effects of Deregulated DNA Damage Signalling on Cancer Chemotherapy Response and Resistance. Nat. Rev. 2012, 12, 587–598. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, X.; Li, D.; Zhu, C.; Guo, X.; Feng, L.; Yu, Z. Biomedicine & Pharmacotherapy PARP Inhibitors in Small Cell Lung Cancer: The Underlying Mechanisms and Clinical Implications. Biomed. Pharmacother. 2022, 153, 113458. [Google Scholar] [CrossRef]
- Sabari, J.K.; Lok, B.H.; Laird, J.H.; Poirier, J.T.; Charles, M.; Sloan, M.; Cancer, K.; Sloan, M.; Cancer, K.; Program, P.; et al. Unravelling the Biology of SCLC: Implications for Therapy. Nat. Rev. Clin. Oncol. 2018, 14, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Bunn, P.A.; Minna, J.D. Small-Cell Lung Cancer: What We Know, What We Need to Know and the Path Forward. Nat. Publ. Gr. 2017, 17, 725–737. [Google Scholar] [CrossRef]
- Foy, V.; Schenk, M.W.; Baker, K.; Gomes, F.; Lallo, A.; Frese, K.K.; Forster, M.; Dive, C.; Blackhall, F. Lung Cancer Targeting DNA Damage in SCLC. Lung Cancer 2017, 114, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, T.; Tong, P.; Diao, L.; Li, L.; Fan, Y.; Hoff, J.; John, V.; Wang, J.; Byers, L.A.; Oncology, N.M.; et al. Targeting AXL and MTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung Cancer. Clin. Cancer Res. 2018, 23, 6239–6253. [Google Scholar] [CrossRef] [Green Version]
- Cardnell, R.J.; Feng, Y.; Mukherjee, S.; Diao, L.; Tong, P.; Stewart, A.; Masrorpour, F.; Fan, Y.; Nilsson, M.; Shen, Y.; et al. Activation of the PI3K / MTOR Pathway Following PARP Inhibition in Small Cell Lung Cancer. PLoS ONE 2016, 1, e0152584. [Google Scholar] [CrossRef]
- Cardnell, J.; Feng, Y.; Diao, L.; Fan, Y.; Masrorpour, F. Proteomic Markers of DNA Repair and PI3K Pathway Activation Predict Response to the PARP Inhibitor BMN 673 in Small Cell Lung Cancer. Clin. Cancer Res. 2014, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; Shen, L.; Liu, W.; Duchemann, B.; Tumula, P.; Girard, L.; et al. Proteomic Profiling Identifies Dysregulated Pathways in Small Cell Lung Cancer and Novel Therapeutic Targets Including PARP1. Cancer Discov. 2013, 2, 798–811. [Google Scholar] [CrossRef]
- Sen, T.; Tong, P.; Stewart, C.A.; Cristea, S.; Valliani, A.; David, S.; Redwood, A.B.; Fan, Y.H.; Li, L.; Glisson, B.S.; et al. CHK1 Inhibition in Small-Cell Lung Cancer Produces SingleAgent Activity in Biomarker-Defined Disease Subsets and Combination Activity with Cisplatin or Olaparib. Cancer Res. 2017, 77, 3870–3884. [Google Scholar] [CrossRef] [Green Version]
- De Bono, J.; Ramanathan, R.K.; Mina, L.; Chugh, R.; Glaspy, J.; Rafii, S.; Kaye, S.; Sachdev, J.; Heymach, J.; Smith, D.C.; et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2018, 7, 620–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, T.; Gay, C.M.; Byers, L.A. Targeting DNA Damage Repair in Small Cell Lung Cancer and the Biomarker Landscape. Transl. Lung Cancer Res. 2018, 7, 50–68. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Giaccone, G. NewS & VIewS Opportunities and Challenges. Nat. Rev. Clin. Oncol. 2018, 15, 348–349. [Google Scholar] [CrossRef]
- Rudin, C.M.; Brambilla, E.; Faivre-finn, C.; Sage, J.; Sloan, M.; Cancer, K.; Sloan, M.; Cancer, K. Small-Cell Lung Cancer Charles. Nat. Rev. Dis. Prim. 2021, 7, 1–43. [Google Scholar] [CrossRef]
- Pavan, A.; Attili, I.; Pasello, G.; Guarneri, V.; Conte, P.F.; Bonanno, L. Immunotherapy in Small-Cell Lung Cancer: From Molecular Promises to Clinical Challenges. J. Immunother. Cancer 2019, 1, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atr, P.K.; Bai, R.; Xie, T.; Ye, X. Recent Advances in Synergistic Antitumor Effects Exploited from the Inhibition of Ataxia Telangiectasia and RAD3-Related. Molecules 2022, 27, 2491. [Google Scholar]
- Van Gent, D.C.; Kanaar, R. Exploiting DNA Repair Defects for Novel Cancer Therapies. Mol. Biol. Cell 2016, 27, 2145–2148. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Carrigan, B.O.; Jackson, S.P.; Yap, T.A. Europe PMC Funders Group Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2017, 7, 20–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Qin, Q.; Liu, B.; Li, X.; Zeng, L.; Wang, J.; Kong, L.; Zhong, D. Targeting DNA-PK Overcomes Acquired Resistance to Third-Generation EGFR-TKI Osimertinib in Non-Small-Cell Lung Cancer. Acta Pharmacol. Sin. 2021, 42, 648–654. [Google Scholar] [CrossRef]
- Pietanza, M.C.; Byers, L.A.; Minna, J.D.; Rudin, C.M. Small Cell Lung Cancer: Will Recent Progress Lead to Improved. Clin. Cancer Res. 2016, 21, 2244–2255. [Google Scholar] [CrossRef]
- Barnieh, F.M.; Loadman, P.M.; Falconer, R.A. Current Research in Pharmacology and Drug Discovery Progress towards a Clinically-Successful ATR Inhibitor for Cancer Therapy. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100017. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Lok, B.H.; Gardner, E.E.; Schneeberger, V.E.; Ni, A.; Desmeules, P.; Rekhtman, N.; De Stanchina, E.; Teicher, B.A.; Riaz, N.; Powell, S.N.; et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin. Cancer Res. 2018, 23, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.E. The Contribution of DNA Repair Pathways to Genome. FEMS Microbiol. Rev. 2022, fuac035, 1–21. [Google Scholar] [CrossRef]
- Sullivan, M.R.; Bernstein, K.A. RAD-ical New Insights into RAD51 Regulation. Genes 2018, 9, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Kooij, B.; Kruswick, A.; van Attikum, H.; Yaffe, M.B. Multi-Pathway DNA-Repair Reporters Reveal Competition between End-Joining, Single-Strand Annealing and Homologous Recombination at Cas9-Induced DNA Double-Strand Breaks. Nat. Commun. 2022, 13, 5295. [Google Scholar] [CrossRef] [PubMed]
- Reginato, G.; Cejka, P. The MRE11 Complex: A Versatile Toolkit for the Repair of Broken DNA. DNA Repai 2020, 91–92, 102869. [Google Scholar] [CrossRef]
- Borgmann, K.; Dornreiter, I.; Petersen, C.; Dikomey, E.; Mansour, W.Y. Involvement of ATM in Homologous Recombination after End Resection and RAD51 Nucleofilament. Nucleic Acids Res. 2015, 43, 3154–3166. [Google Scholar] [CrossRef] [Green Version]
- Beinse, G.; Just, P.; Belda, M.L.F.; Laurent-puig, P.; Jacques, S.; Koual, M.; Garinet, S.; Leroy, K.; Delanoy, N.; Blons, H.; et al. Discovery and Validation of a Transcriptional Signature Identifying Homologous Recombination-de Fi Cient Breast, Endometrial and Ovarian Cancers. Br. J. Cancer 2022, 127, 1123–1132. [Google Scholar] [CrossRef]
- Ji, W.; Weng, X.; Xu, D.; Cai, S.; Lou, H.; Ding, L. Biochemical and Biophysical Research Communications Non-Small Cell Lung Cancer Cells with de Fi Ciencies in Homologous Recombination Genes Are Sensitive to PARP Inhibitors. Biochem. Biophys. Res. Commun. 2020, 522, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; Mclellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; Mcmichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Pishvaian, M.J.; Brody, J.R.; Chen, W.; Marshall, J.L. Prevalence of Homologous Recombination—Related Gene Mutations Across Multiple Cancer Types Abstract. JCO Precis. Oncol. 2018, 2018, PO.17.00286. [Google Scholar]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Gregory, P. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diossy, M.; Borcsok, J.; Krzystanek, M.; Tisza, V.; Spisak, S.; Rusz, O. OPEN A Subset of Lung Cancer Cases Shows Robust Signs of Homologous Recombination de Fi Ciency Associated Genomic Mutational Signatures. NPJ Precis. Oncol. 2021, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Kee, Y.; Huang, T. Role of Deubiquitinating Enzymes in DNA Repair. Mol. Cell. Biol. 2016, 36, 524–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.T.; Goldberg, A.L. The Deubiquitinating Enzyme Usp14 Allosterically Inhibits Multiple Proteasomal Activities and Ubiquitin-Independent Proteolysis. J. Biol. Chem. 2017, 292, 9830–9839. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.M.; Yang, S.C.; Jo, H.J.; Song, S.Y.; Jeon, Y.J.; Jang, T.W.; Kim, D.J.; Jang, S.H.; Yang, S.H.; Kim, Y.D.; et al. Proteins Involved in DNA Damage Response Pathways and Survival of Stage I Non-Small-Cell Lung Cancer Patients. Ann. Oncol. 2012, 23, 2088–2093. [Google Scholar] [CrossRef]
- Yin, M.; Liao, Z.; Huang, Y.; Liu, Z.; Yuan, X.; Gomez, D. Polymorphisms of Homologous Recombination Genes and Clinical Outcomes of Non-Small Cell Lung Cancer Patients Treated with Definitive Radiotherapy. PLoS ONE 2011, 6, e20055. [Google Scholar] [CrossRef] [Green Version]
- Seedhouse, C.; Faulkner, R.; Ashraf, N.; Das-gupta, E. Polymorphisms in Genes Involved in Homologous Recombination Repair Interact to Increase the Risk of Developing Acute Myeloid Leukemia. Clin. Cancer Res. 2004, 10, 2675–2680. [Google Scholar] [CrossRef] [Green Version]
- Remon, J.; Besse, B.; Leary, A.; Bièche, I.; Job, B.; Lacroix, L.; Auguste, A.; Mauduit, M.; Audigier-valette, C.; Raimbourg, J.; et al. Somatic and Germline BRCA 1 and 2 Mutations in Advanced NSCLC From the SAFIR02-Lung Trial. JTO Clin. Res. Rep. 2020, 1, 100068. [Google Scholar] [CrossRef] [PubMed]
- Lok, B.H.; Rudin, C.M. Epigenetic Targeting of DNA Repair in Lung Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 22429–22431. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Almasan, A. USP14 Regulates DNA Damage Response and Is a Target for Radiosensitization in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2020, 21, 6383. [Google Scholar] [CrossRef]
- Feng, M.; Wang, Y.; Wei, J.M.G. CRL4A DTL Degrades DNA-PKcs to Modulate NHEJ Repair and Induce Genomic Instability and Subsequent Malignant Transformation. Oncogene 2021, 40, 2096–2111. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Hromas, R.; Lee, S.-U. New Research Directions in DNA Repair—Emerging Features of DNA Double-Strand Break Repair in Humans; Chen, C., Ed.; IntechOpen: London, UK, 2013; Chapter 7. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wang, X.Q. XLF-Mediated NHEJ Activity in Hepatocellular Carcinoma Therapy Resistance. BMC Cancer 2017, 17, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, M.; Liao, Z.; Liu, Z.; Wang, L.E.; O’Reilly, M.; Gomez, D.; Li, M.; Komaki, R.; Wei, Q. Genetic Variants of the Nonhomologous End Joining Gene LIG4 and Severe Radiation Pneumonitis in Nonsmall Cell Lung Cancer Patients Treated with Definitive Radiotherapy. Cancer 2012, 118, 528–535. [Google Scholar] [CrossRef] [Green Version]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers 2017, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Sears, C.R.; Sears, C.R. DNA repair as an emerging target for COPD-lung cancer overlap. Respir. Investig. 2019, 57, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Ottenheijm, C.A.C.; Heunks, L.M.A.; Dekhuijzen, R.P.N. Diaphragm adaptations in patients with COPD. Respir. Res. 2008, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Gol, T.M.; Rodemann, H.P.; Dittmann, K. Depletion of Akt1 and Akt2 Impairs the Repair of Radiation-Induced DNA Double Strand Breaks via Homologous Recombination. Int. J. Mol. Sci. 2019, 20, 6316. [Google Scholar]
- Toulany, M.; Iida, M.; Lettau, K.; Coan, J.P.; Rebholz, S.; Khozooei, S.; Harari, P.M.; Wheeler, D.L. Targeting HER3-Dependent Activation of Nuclear AKT Improves Radiotherapy of Non-Small Cell Lung Cancer. Radiother. Oncol. 2022, 174, 92–100. [Google Scholar] [CrossRef]
- Weeden, C.E.; Chen, Y.; Ma, S.B.; Hu, Y.; Ramm, G.; Sutherland, K.D.; Smyth, G.K. Lung Basal Stem Cells Rapidly Repair DNA Damage Using the Error-Prone Nonhomologous End-Joining Pathway. PLOS Biol. 2017, 15, e2000731. [Google Scholar] [CrossRef] [PubMed]
- Meas, R.; Wyrick, J.J.; Smerdon, M.J. Nucleosomes Regulate Base Excision Repair in Chromatin. Mutat. Res. Mol. Mech. Mutagen. 2019, 780, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Klapacz, J.; Pottenger, L.H.; Engelward, B.P.; Heinen, C.D.; Johnson, G.E.; Clewell, R.A.; Carmichael, P.L.; Adeleye, Y.; Andersen, M.E. Contributions of DNA Repair and Damage Response Pathways to the Non-Linear Genotoxic Responses of Alkylating Agents. Mutat. Res. Mol. Mech. Mutagen. 2017, 767, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Manuscript, A.; Repair, B.E. Base Excision Repair and Cancer. Cancer Lett. 2013, 327, 73–89. [Google Scholar] [CrossRef] [Green Version]
- Grundy, G.J.; Parsons, J.L. Base Excision Repair and Its Implications to Cancer Therapy. Essays Biochem. 2020, 64, 831–843. [Google Scholar] [PubMed]
- Yousafzai, N.A.; Zhou, Q.; Xu, W.; Shi, Q.; Xu, J.; Feng, L.; Jin, H.; Wang, X.; Chen, H.; Shin, V.Y. SIRT1 Deacetylated and Stabilized XRCC1 to Promote Chemoresistance in Lung Cancer. Cell Death Dis. 2019, 10, 363. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xu, C.; Chen, H.; Jia, J.; Wang, L.; Feng, H.; Wang, H.; Song, Z.; Yang, N.; Zhang, Y. Lung Cancer Genomic Alterations and Clinical Outcomes in Patients with Lung Adenocarcinoma with Transformation to Small Cell Lung Cancer after Treatment with EGFR Tyrosine Kinase Inhibitors: A Multicenter Retrospective Study. Lung Cancer 2021, 155, 20–27. [Google Scholar] [CrossRef]
- Inhibitors, R.K. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells 2018, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Cai, X.; Li, J. Histological Transformation of Lung Adenocarcinoma to Small Cell Lung Cancer with Mutant C797S Conferring Acquired Resistance to Osimertinib. J. Int. Med. Res. 2020, 48, 1–7. [Google Scholar] [CrossRef]
- Insa, A.; Majem, M.; Isla, D.; Costa, E.C.; Puig, M.; Kraemer, S.; Schnell, D. A Phase Ib Trial of Continuous Once-Daily Oral Afatinib plus Sirolimus in Patients with Epidermal Growth Factor Receptor Mutation-Positive Non-Small Cell Lung Cancer and/or Disease Progression Following Prior Erlotinib or Gefitinib. Lung Cancer 2017, 108, 154–160. [Google Scholar] [CrossRef]
- Mok, T.; Nakagawa, K.; Rosell, R.; Lee, K.; Corral, J.; Migliorino, M.; Pluzanski, A.; Linke, R.; Devgan, G.; Sbar, E.; et al. MA26.11 Effects of Dose Modifications on the Safety and Efficacy of Dacomitinib for EGFR Mutation-Positive NSCLC. J. Thorac. Oncol. 2018, 13, S454. [Google Scholar] [CrossRef]
- Wang, W.; Xu, J.; Chong, J.; Wang, D. Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair. DNA Repair 2018, 71, 43–55. [Google Scholar] [CrossRef]
- Shah, G.M. Methods to Study Intracellular Movement and Localization of the Nucleotide Excision Repair Proteins at the DNA Lesions in Mammalian. Front. Cell Dev. Biol. 2020, 8, 590242. [Google Scholar] [CrossRef]
- Id, K.S.; Id, D.A.P.; Id, P.M.; Id, J.J.W. Set2 Histone Methyltransferase Regulates Transcription Coupled-Nucleotide Excision Repair in Yeast. PLoS Genet. 2022, 18, e1010085. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B. Pharmacogenetics of Platinum-Based Chemotherapy in Non-Small Cell Lung Cancer: Predictive Validity of Polymorphisms of ERCC1. Expert Opin. Drug Metab. Toxicol. 2018, 14, 1744–7607. [Google Scholar] [CrossRef] [PubMed]
- Pilji, M.; Mladinov, S.; Matana, A.; Kuret, S. Low ERCC1 Expression Is a Good Predictive Marker in Lung Adenocarcinoma Patients Receiving Chemotherapy Based on Platinum in All TNM Stages—A Single-Center Study. Diagn. Pathol. 2019, 14, 105. [Google Scholar]
- Miyake, N.; Chikumi, H.; Yamaguchi, K.; Takata, M.; Takata, M.; Okada, K. Effect of Cetuximab and EGFR Small Interfering RNA Combination Treatment in NSCLC Cell Lines with Wild Type EGFR and Use of KRAS as a Possible Biomarker for Treatment Responsiveness. Yonago Acta Med. 2019, 62, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Shen, M.; Han, N.; Xu, H.; Peng, X. Photodiagnosis and Photodynamic Therapy Chlorin E6 Mediated Photodynamic Therapy Triggers Resistance through ATM-Related DNA Damage Response in Lung Cancer Cells. Photodiagnosis Photodyn. Ther. 2022, 37, 102645. [Google Scholar] [CrossRef]
- Kaur, E.; Ketkar, M.; Dutt, S. Glioblastoma Recurrent Cells Switch between ATM and ATR Pathway as an Alternative Strategy to Survive Radiation Stress. Med. Oncol. 2022, 39, 50. [Google Scholar] [CrossRef]
- Esposito, F.; Giuffrida, R.; Raciti, G.; Puglisi, C.; Forte, S. Wee1 Kinase: A Potential Target to Overcome Tumor Resistance to Therapy. Int. J. Mol. Sci. 2021, 22, 10689. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhao, H.; Li, C.; Li, P. An MTOR and DNA-PK Dual Inhibitor CC-115 Hinders Non-Small Cell Lung Cancer Cell Growth. Cell Death Discov. 2022, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, A.B.; Chan, G.K.; Gamper, A.M. Targeting the DNA Damage Response for Cancer Therapy by Inhibiting the Kinase Wee1. Front. Oncol. 2022, 12, 828684. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Zhao, Y.; Arkenau, H.; Lao, T.; Chu, L.; Xu, Q. Signal Pathways and Precision Therapy of Small-Cell Lung Cancer. Signal Transduct. Target. Ther. 2022, 7, 187. [Google Scholar] [CrossRef]
- Knelson, E.H.; Patel, S.A. PARP Inhibitors in Small-Cell Lung Cancer: Rational Combinations to Improve Responses. Cancers 2021, 13, 727. [Google Scholar] [CrossRef] [PubMed]
- Groelly, F.J.; Tarsounas, M. Review DNA Damage and Cancer Immunotherapy: A STING in the Tale. Mol. Cell 2020, 80, 21–28. [Google Scholar] [CrossRef]
- Lee, H.; Paull, T.T. Cellular Functions of the Protein Kinase ATM and Their Relevance to Human Disease. Mol. Cell Biol. 2021, 22, 796–814. [Google Scholar] [CrossRef]
- Ghosh, S.; Ghosh, A. Activation of DNA Damage Response Signaling in Mammalian Cells by Ionizing Radiation. Free Radic. Res. 2021, 55, 814–827. [Google Scholar] [CrossRef]
- Simoneau, A.; Zou, L. ScienceDirect An Extending ATR–CHK1 Circuitry: The Replication Stress Response and Beyond. Curr. Opin. Genet. Dev. 2021, 71, 92–98. [Google Scholar] [CrossRef]
- Rajapakse, V.N.; Pommier, Y.; Khan, J.; Thomas, C.J. Article Therapeutic Targeting of ATR Yields Durable Regressions in Small Cell Lung Cancers with High Replication Stress Therapeutic Targeting of ATR Yields Durable Regressions in Small Cell Lung Cancers with High Replication Stress. Cancer Cell 2021, 39, 566–579.e7. [Google Scholar] [CrossRef]
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of Eukaryotic Topoisomerases in Transcription, Replication and Genomic Stability. Nat. Rev. Mol. Cell Biol. 2022, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Dunne, V.; Ghita, M.; Small, D.M.; Coffey, C.B.M.; Weldon, S.; Taggart, C.C.; Osman, S.O.; Mcgarry, C.K.; Prise, K.M.; Hanna, G.G.; et al. Inhibition of Ataxia Telangiectasia Related-3 ( ATR ) Improves Therapeutic Index in Preclinical Models of Non-Small Cell Lung Cancer ( NSCLC ) Radiotherapy. Radiother. Oncol. 2017, 124, 475–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, A.; Redon, C.E.; Sciuto, L.; Padiernos, E.; Ji, J.; Lee, M.; Yuno, A.; Lee, S.; Zhang, Y.; Tran, L.; et al. Phase I Study of ATR Inhibitor M6620 in Combination With Topotecan in Patients With Advanced Solid Tumors. J. Clin. Oncol. 2018, 36, 1594–1602. [Google Scholar] [CrossRef] [PubMed]
- Jo, U.; Senatorov, I.S.; Zimmermann, A.; Saha, L.K.; Murai, Y.; Kim, S.H.; Rajapakse, V.N.; Elloumi, F.; Takahashi, N.; Schultz, C.W.; et al. Novel and Highly Potent ATR Inhibitor M4344 Kills Cancer Cells With Replication Stress, and Enhances the Chemotherapeutic Activity of Widely Used DNA Damaging Agents. Mol. Cancer Ther. 2021, 20, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.; Ghosh, M.; Kim, T.H.; Park, N.; Pandey, K.; Cho, Y.B.; Hong, S.D.; Katuwal, N.B.; Kang, M.; An, H.J.; et al. Synergism of AZD6738, an ATR Inhibitor, in Combination with Belotecan, a Camptothecin Analogue, in Chemotherapy-Resistant Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 1223. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, K.; Stewart, C.A.; Cargill, K.R.; Della Corte, C.M.; Wang, Q.; Shen, L.; Diao, L.; Cardnell, R.J.; Peng, D.H.; Rodriguez, B.L.; et al. AXL Inhibition Induces DNA Damage and Replication Stress in Non-Small Cell Lung Cancer Cells and Promotes Sensitivity to ATR Inhibitors. Mol. Cancer Res. 2021, 19, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Wis, H.C.; Iy, G.V.; Moo, K.; Sarah, M.T. Activity of M3814, an Oral DNA-PK Inhibitor, In Combination with Topoisomerase II Inhibitors in Ovarian Cancer Models. Sci. Rep. 2019, 9, 18882. [Google Scholar] [CrossRef] [Green Version]
- Harnor, S.J.; Brennan, A.; Cano, C.Ø. Targeting DNA-Dependent Protein Kinase for Cancer Therapy. ChemMedChem 2017, 2017, 895–900. [Google Scholar] [CrossRef] [Green Version]
- Timme, C.R.; Rath, B.H.; Neill, J.W.O.; Camphausen, K.; Tofilon, P.J. The DNA-PK Inhibitor VX-984 Enhances the Radiosensitivity of Glioblastoma Cells Grown In Vitro and as Orthotopic Xenografts. Mol. Cancer Ther. 2019, 17, 1207–1216. [Google Scholar] [CrossRef] [Green Version]
- Zenke, F.T.; Zimmermann, A.; Sirrenberg, C.; Dahmen, H.; Kirkin, V.; Pehl, U.; Grombacher, T.; Wilm, C.; Fuchss, T.; Amendt, C.; et al. Pharmacologic Inhibitor of DNA-PK, M3814, Potentiates Radiotherapy and Regresses Human Tumors in Mouse Models. Mol. Cancer Ther. 2020, 19, 1091–1101. [Google Scholar] [CrossRef] [Green Version]
- Carr, M.I.; Zimmermann, A.; Chiu, L.; Zenke, F.T.; Blaukat, A.; Vassilev, L.T. DNA-PK Inhibitor, M3814, as a New Combination Partner of Mylotarg in the Treatment of Acute Myeloid Leukemia. Front. Oncol. 2020, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhu, M.; Shen, W.; Wang, C.; Dai, J.; Xu, L.; Jin, G.; Hu, Z.; Ma, H.; Shen, H. ORIGINAL ARTICLE A Potentially Functional Polymorphism in ABCG2 Predicts Clinical Outcome of Non-Small Cell Lung Cancer in a Chinese Population. Pharm. J. 2017, 17, 280–285. [Google Scholar] [CrossRef]
- Zou, C.; Chen, Z. M3814, a DNA-PK Inhibitor, Modulates ABCG2-Mediated Multidrug Resistance in Lung Cancer. Front. Oncol. 2020, 10, 674. [Google Scholar] [CrossRef]
- Vestergaard, H.H.; Christensen, M.R.; Lassen, U.N.; Vestergaard, H.H.; Christensen, M.R.; Lassen, U.N. A Systematic Review of Targeted Agents for Non-Small Cell Lung Cancer. Acta Oncol. 2018, 57, 176–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fumarola, C.; Bonelli, M.A.; Petronini, P.G.; Alfieri, R.R. Targeting PI3K / AKT / MTOR Pathway in Non Small Cell Lung Cancer. Biochem. Pharmacol. 2014, 90, 197–207. [Google Scholar] [CrossRef]
- Tan, A.C. Targeting the PI3K / Akt / MTOR Pathway in Non-Small Cell Lung Cancer ( NSCLC ). Thorac. Cancer 2020, 11, 511–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, B.; Sun, X.; Chen, X.; Chen, Z.; Zhu, W.; Zhu, H.; Gu, D. Dual Inhibition of DNA-PKcs and MTOR by CC-115 Potently Inhibits Human Renal Cell Carcinoma Cell Growth. Aging 2020, 12, 20445–20456. [Google Scholar] [CrossRef]
- Bürkel, F.; Jost, T.; Hecht, M.; Heinzerling, L.; Fietkau, R.; Distel, L. Dual MTOR/DNA-PK Inhibitor CC-115 Induces Cell Death in Melanoma Cells and Has Radiosensitizing Potential. Int. J. Mol. Sci. 2020, 21, 9321. [Google Scholar] [CrossRef] [PubMed]
- Munster, P.; Mita, M.; Mahipal, A.; Nemunaitis, J.; Massard, C.; Mikkelsen, T.; Cruz, C.; Paz-ares, L.; Hidalgo, M.; Rathkopf, D.; et al. First-In-Human Phase I Study Of A Dual MTOR Kinase And DNA-PK Inhibitor (CC-115) In Advanced Malignancy. Cancer Manag. Res. 2019, 11, 10463–10476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, T.; Sapinoso, L.M.; Tran, T.; Gaffney, B.; Wong, L.; Raymon, H.K.; Mortensen, D.S.; Xu, S. CC-115, a Dual Inhibitor of MTOR Kinase and DNA-PK, Blocks DNA Damage Repair Pathways and Selectively Inhibits ATM-Deficient Cell Growth in Vitro. Oncotarget 2017, 8, 74688–74702. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Chen, S.; Wei, Y.; Wei, X. DNA-PK Inhibition by M3814 Enhances Chemosensitivity in Non-Small Cell Lung Cancer. Acta Pharm. Sin. B 2021, 11, 3935–3949. [Google Scholar] [CrossRef]
- Saar, M.; Narits, J.; Mägi, L.; Aaspõllu, H.; Vapper, A. Expression of Immune Checkpoint PD-1 in Non-Small Cell Lung Cancer Is Associated with Tumor Cell DNA—Dependent Protein Kinase. Mol. Clin. Oncol. 2021, 15, 211. [Google Scholar] [CrossRef]
- Ashworth, A.; Lord, C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018, 15, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Fugger, K.; Bajrami, I.; Silva, M.; Santos, D.; Young, S.J.; Kelly, G.; Hewitt, G.; Patel, H.; Goldstone, R.; Boulton, S.J.; et al. Europe PMC Funders Group Targeting the Nucleotide Salvage Factor DNPH1 Sensitizes BRCA—Deficient Cells to PARP Inhibitors. Science 2021, 372, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Nan, A.; Chen, L.; Zhang, N.; Jia, Y.; Li, X.; Zhou, H.; Ling, Y. Circular RNA CircNOL10 Inhibits Lung Cancer Development by Promoting SCLM1-Mediated Transcriptional Regulation of the Humanin Polypeptide Family. Adv. Sci. 2019, 6, 1800654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, M.; Mukhopadhyay, A.; Sharawat, S.K.; Kumar, S. BBA—Reviews on Cancer Role of MicroRNAs in Regulating Cell Proliferation, Metastasis and Chemoresistance and Their Applications as Cancer Biomarkers in Small Cell Lung Cancer. BBA Rev. Cancer 2021, 1876, 188552. [Google Scholar] [CrossRef]
- Carlisle, J.W. An Update on the Immune Landscape in Lung and Head and Neck Cancers. CA Cancer J. Clin. 2020, 70, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Drapkin, B.J.; Farago, A.F. Unexpected Synergy Reveals New Therapeutic Strategy in SCLC. Trends Pharmacol. Sci. 2019, 40, 295–297. [Google Scholar] [CrossRef]
- Jin, R.; Liu, B.; Yu, M.; Song, L.; Gu, M.; Wang, Z.; Li, X.; Zhang, X.; Wang, J.; Ma, T. Profiling of DNA damage and repair pathways in small cell lung cancer reveals a suppressive role in the immune landscape. Mol. Cancer 2021, 20, 130. [Google Scholar] [CrossRef] [PubMed]
- Owonikoko, T.K.; Dahlberg, S.E.; Sica, G.L.; Wagner, L.I. Original Report Abstract Randomized Phase II Trial of Cisplatin and Etoposide in Combination With Veliparib or Placebo for Extensive-Stage Small-Cell Lung Cancer: ECOG-ACRIN 2511 Study. J. Clin. Oncol. 2019, 37, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Yeap, B.Y.; Stanzione, M.; Hung, Y.P.; Rebecca, S.; Marcoux, J.P.; Zhong, J.; Rangachari, D.; Barbie, D.A.; Myers, D.T.; et al. Combination Olaparib and Temozolomide in Relapsed Small Cell Lung Cancer. Cancer Discov. 2020, 9, 1372–1387. [Google Scholar] [CrossRef] [Green Version]
- Lines, C.C. Talazoparib Is a Potent Radiosensitizer in Small Cell Lung Cancer Cell Lines and Xenografts. Clin. Cancer Res. 2019, 24, 5143–5152. [Google Scholar] [CrossRef] [Green Version]
- Sibanda, B.L.; Chirgadze, D.Y.; Ascher, D.B. DNA-PKcs Structure Suggests an Allosteric Mechanism Modulating DNA Double-Strand Break Repair. Science 2017, 194, 520–524. [Google Scholar] [CrossRef]
- Genet, N. Integrative Genome Analyses Identify Key Somatic Driver Mutations of Small Cell Lung Cancer. Nat. Genet. 2016, 44, 1104–1110. [Google Scholar] [CrossRef]
- Fujimoto, J.; Cristea, S.; Nguyen, T.; Diao, L.; Fan, Y.; Yang, Y.; Wang, J.; Glisson, B.S.; Wistuba, I.I.; Sage, J.; et al. Targeting DNA Damage Response Promotes Anti-Tumor Immunity through STING-Mediated T-Cell Activation in Small Cell Lung Cancer. Cancer Discov. 2020, 9, 646–661. [Google Scholar] [CrossRef] [Green Version]
- Pietanza, M.C.; Kadota, K.; Huberman, K.; Sima, C.S.; Fiore, J.J.; Sumner, D.K.; Travis, W.D.; Heguy, A.; Ginsberg, M.S.; Holodny, A.I.; et al. Phase II Trial of Temozolomide in Patients with Relapsed Sensitive or Refractory Small Cell Lung Cancer, with Assessment of Methylguanine-DNA Methyltransferase as a Potential Biomarker. Clin. Cancer Res. 2012, 18, 1138–1145. [Google Scholar] [CrossRef] [Green Version]
- Reichert, Z.R.; Wahl, D.R.; Maybaum, J.; Connor, M.J.O. PARP1 Trapping and DNA Replication Stress Enhance Radiosensitization with Combined WEE1 and PARP Inhibitors. Mol. Cancer Res. 2019, 16, 222–232. [Google Scholar] [CrossRef]
- Hiddinga, B.I.; Raskin, J.; Janssens, A.; Pauwels, P.; Van Meerbeeck, J.P. Recent developments in the treatment of small cell lung cancer. Eur. Respir. Rev. 2021, 30, 210079. [Google Scholar] [CrossRef]

| ID | Condition or Disease | Intervention/Treatment | Status |
|---|---|---|---|
| NCT02487095 | Carcinoma, Non-small cell lung Ovarian neoplasms Small cell lung carcinoma Uterine cervical neoplasms carcinoma, Neuro-endocrine Extra-pulmonary Small cell cancer | Drug: Topotecan Drug: VX-970 (M6620) | Active, Not recruiting |
| NCT02487095 | Carcinoma, non-Small cell lung Ovarian neoplasms Small cell lung carcinoma Uterine cervical Neoplasms carcinoma, Neuroendocrine extrapulmonary Small cell cancer | Drug: Topotecan Drug: VX-970 (M6620) | Active, Not recruiting |
| NCT02589522 | Metastatic lung neuroendocrine neoplasm Metastatic lung non-small cell carcinoma Metastatic lung small cell carcinoma Metastatic malignant neoplasm in the brain stage IV lung cancer AJCC v8Stage IVA lung Cancer AJCC v8Stage IVB lung Cancer AJCC v8 | Drug: Berzosertib Other: Quality-of-Life Assessment Procedure: Therapeutic Conventional Surgery Radiation: Whole-Brain Radiotherapy | Active, Not recruiting |
| NCT04768296 | Small cell lung cancer | Drug: Berzosertib Drug: Topotecan | Active, Not recruiting |
| NCT04216316 | Lung non-small cell squamous carcinoma Stage IV lung cancer AJCC v8 | Drug: Berzosertib Procedure: Biospecimen Collection Drug: Carboplatin Drug: Gemcitabine Hydrochloride Biological: Pembrolizumab | Recruiting |
| NCT05450692 | Advanced or metastatic non-small cell lung cancer | Drug: Ceralasertib Drug: Durvalumab Drug: Docetaxel | Not yet recruiting |
| NCT04699838 | Extensive stage small cell lung cancer | Drug: Cisplatin Drug: Carboplatin Drug: Etoposide Drug: Durvalumab Drug: Ceralasertib | Recruiting |
| NCT04768296 | Small cell lung cancer | Drug: Berzosertib Drug: Topotecan | Active, not recruiting |
| NCT04216316 | Lung non-small cell squamous carcinoma Stage IV lung cancer AJCC v8 | Drug: Berzosertib Procedure: Biospecimen Collection Drug: Carboplatin Drug: Gemcitabine Hydrochloride Biological: Pembrolizumab | Recruiting |
| NCT04826341 | HRD cancer SCLC Advanced solid tumors | Drug: Berzosertib Drug: Sacituzumab Govitecan | Recruiting |
| ID | Condition/Disease | Intervention | Status |
|---|---|---|---|
| NCT01638546 | Recurrent small cell lung carcinoma | Other: Laboratory Biomarker Analysis Other: Placebo Drug: Temozolomide Drug: Veliparib | Completed |
| NCT04728230 | Extensive stage lung small cell carcinoma Stage IV lung cancer AJCC v8 Stage IVA lung cancer AJCC v8 Stage IVB lung cancer AJCC v8 | Drug: Carboplatin Biological: Durvalumab Drug: Etoposide Drug: Olaparib Radiation: Radiation Therapy | Recruiting |
| NCT04701307 | Lung small cell carcinoma Neuroendocrine carcinoma Stage III lung cancer AJCC v8 Stage IIIA lung cancer AJCC v8 Stage IIIB lung cancer AJCC v8 Stage IIIC lung cancer AJCC v8 | Biological: Dostarlimab Drug: Niraparib | Recruiting |
| NCT03672773 | Recurrent extensive stage small cell lung carcinoma refractory extensive stage small cell lung carcinoma | Drug: Talazoparib Drug: Temozolomide | Active, Not recruiting |
| NCT02289690 | Small cell lung cancer | Drug: Veliparib Drug: Carboplatin Drug: Etoposide Drug: Placebo | Completed |
| NCT04538378 | EGFR-mutated non-small cell lung carcinoma Small cell/neuroendocrine | Drug: Olaparib Drug: Durvalumab | Recruiting |
| NCT03830918 | Advanced malignant solid neoplasm Extensive stage lung small cell carcinoma Stage III lung cancer AJCC v8 Stage IIIA lung cancer AJCC v8 Stage IIIB lung cancer AJCC v8 Stage IIIC lung cancer AJCC v8 Stage IV lung cancer AJCC v8 Stage IVA lung cancer AJCC v8 Stage IVB lung cancer AJCC v8 | Biological: Atezolizumab Drug: Niraparib Other: Quality-of-Life Assessment Other: Questionnaire Administration Drug: Temozolomide | Recruiting |
| NCT01286987 | Advanced or recurrent solid tumors Breast neoplasms Ovarian cancer, epithelial Ewing sarcoma Small cell lung carcinoma Prostate cancer Pancreas cancer | Drug: Talazoparib | Completed |
| NCT03958045 | Small cell lung cancer | Combination Product: Rucaparib and Nivolumab | Recruiting |
| NCT02769962 | Urothelial carcinoma Urothelial cancer Lung neoplasms Small cell lung cancer Prostate cancer | Drug: EP0057 Drug: Olaparib | Recruiting |
| NCT01082549 | Squamous cell lung cancer | Drug: gemcitabine/carboplatin Drug: gemcitabine/carboplatin plus Iniparib | Completed |
| NCT01788332 | Non-small cell lung cancer | Drug: Olaparib Other: Placebo | Unknown |
| NCT01086254 | Non-small cell lung cancer stage IV | Drug: Iniparib Drug: gemcitabine Drug: cisplatin | Completed |
| NCT05392686 | Non-small cell lung cancer | Drug: PD-1 inhibitor Drug: PARP inhibitor | Recruiting |
| NCT04380636 | Lung neoplasms Carcinoma, non-small cell lung | Biological: Pembrolizumab Drug: Olaparib Drug: Placebo for Olaparib Drug: Etoposide Drug: Carboplatin Drug: Cisplatin Drug: Paclitaxel Drug: Pemetrexed Radiation: Thoracic Radiotherapy Drug: Durvalumab | Recruiting |
| NCT01560104 | Non-small cell lung cancer | Drug: Veliparib Drug: Carboplatin Drug: Paclitaxel Drug: Placebo | Completed |
| NCT02944396 | Non-small cell lung cancer | Drug: Pemetrexed Drug: Nivolumab Drug: Paclitaxel Drug: Veliparib Drug: Carboplatin | Completed |
| NCT02264990 | Non-squamous non-small cell lung cancer | Drug: Paclitaxel Drug: Carboplatin Drug: Cisplatin Drug: Veliparib Drug: Pemetrexed | Completed |
| NCT04538378 | EGFR-mutated non-small cell lung Carcinoma Small cell/neuroendocrine | Drug: Olaparib Drug: Durvalumab | Recruiting |
| NCT02106546 | Squamous non-small cell lung cancer | Drug: Carboplatin Drug: Veliparib Drug: Paclitaxel Drug: Placebo to veliparib | Completed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venugopala, K.N. Targeting the DNA Damage Response Machinery for Lung Cancer Treatment. Pharmaceuticals 2022, 15, 1475. https://doi.org/10.3390/ph15121475
Venugopala KN. Targeting the DNA Damage Response Machinery for Lung Cancer Treatment. Pharmaceuticals. 2022; 15(12):1475. https://doi.org/10.3390/ph15121475
Chicago/Turabian StyleVenugopala, Katharigatta N. 2022. "Targeting the DNA Damage Response Machinery for Lung Cancer Treatment" Pharmaceuticals 15, no. 12: 1475. https://doi.org/10.3390/ph15121475






