Mitochondrion-Targeted NIR Therapeutic Agent Suppresses Melanoma by Inducing Apoptosis and Cell Cycle Arrest via E2F/Cyclin/CDK Pathway
Abstract
:1. Introduction
2. Results
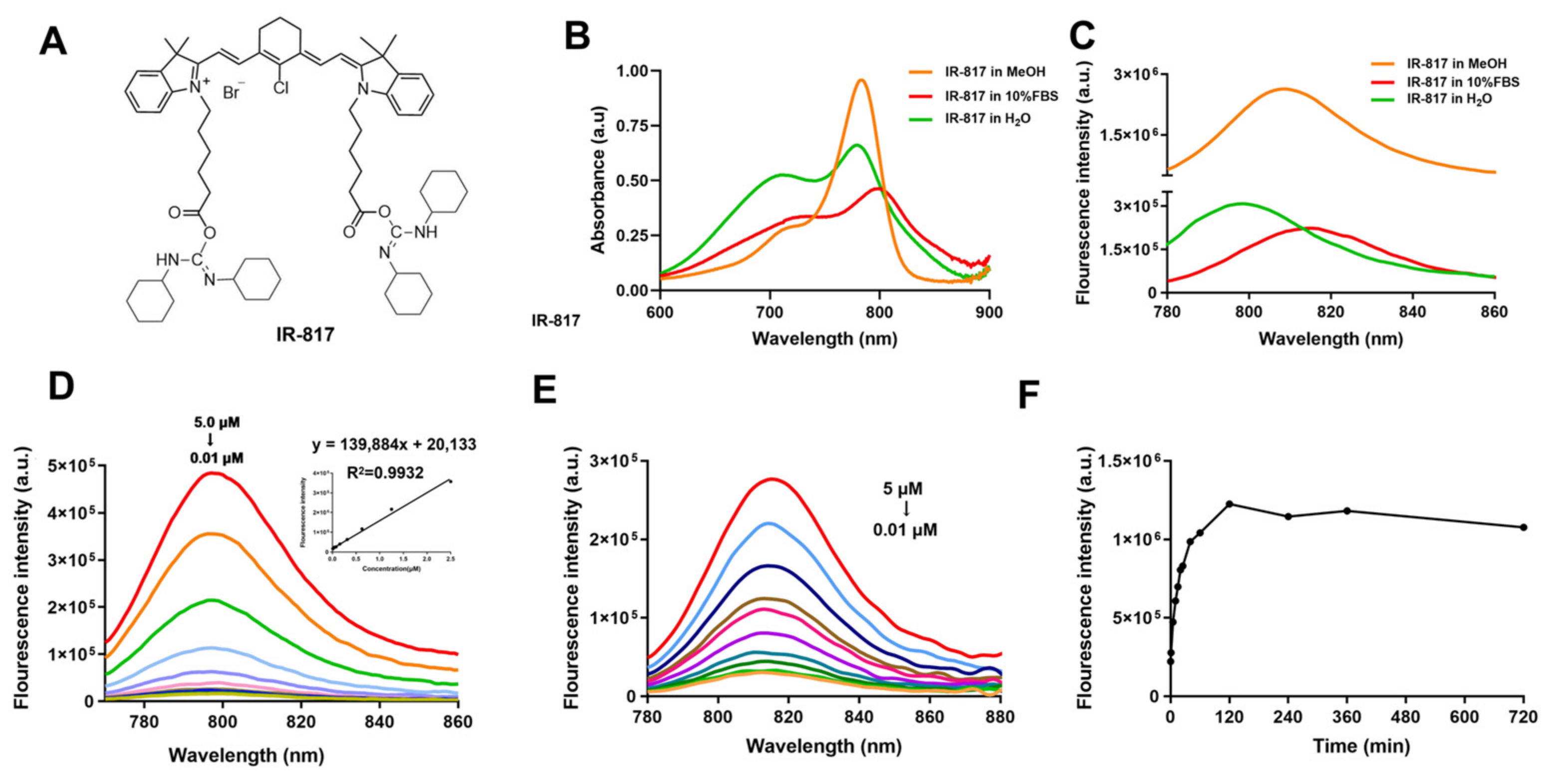
2.1. Chemical Synthesis and Optical Properties of IR-817
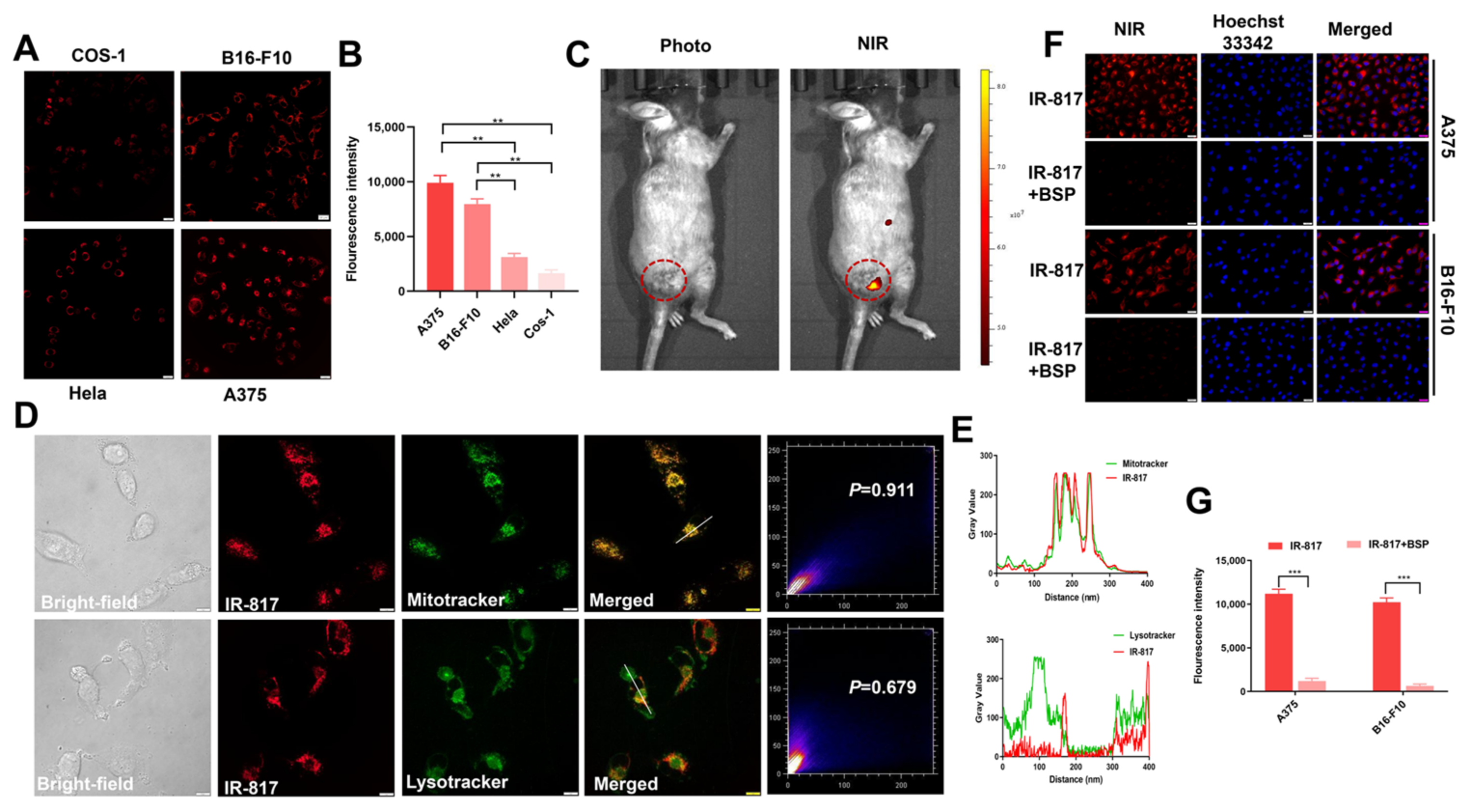
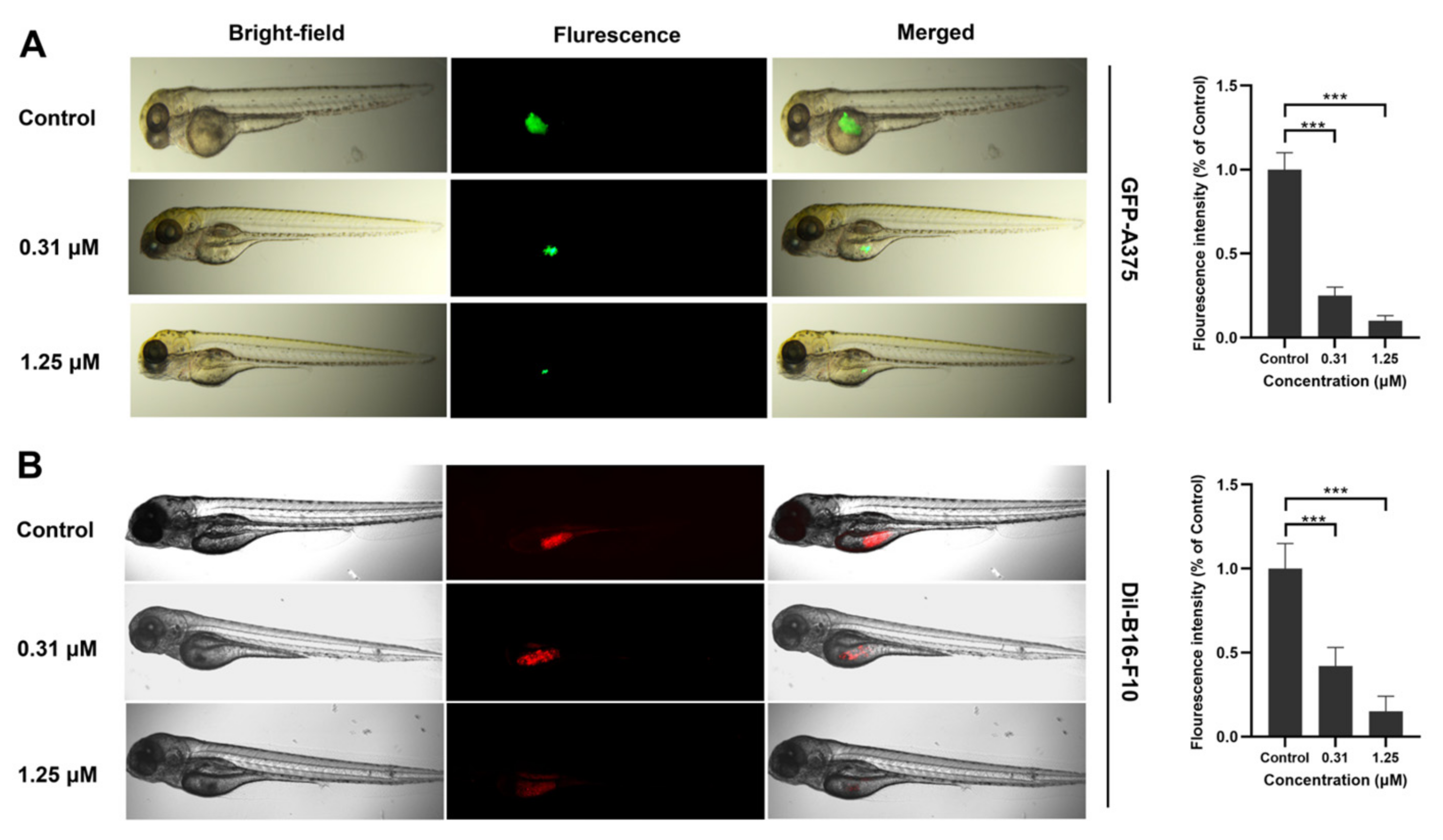
2.2. Identification of IR-817 for Melanoma Targeting Imaging In Vitro and In Vivo
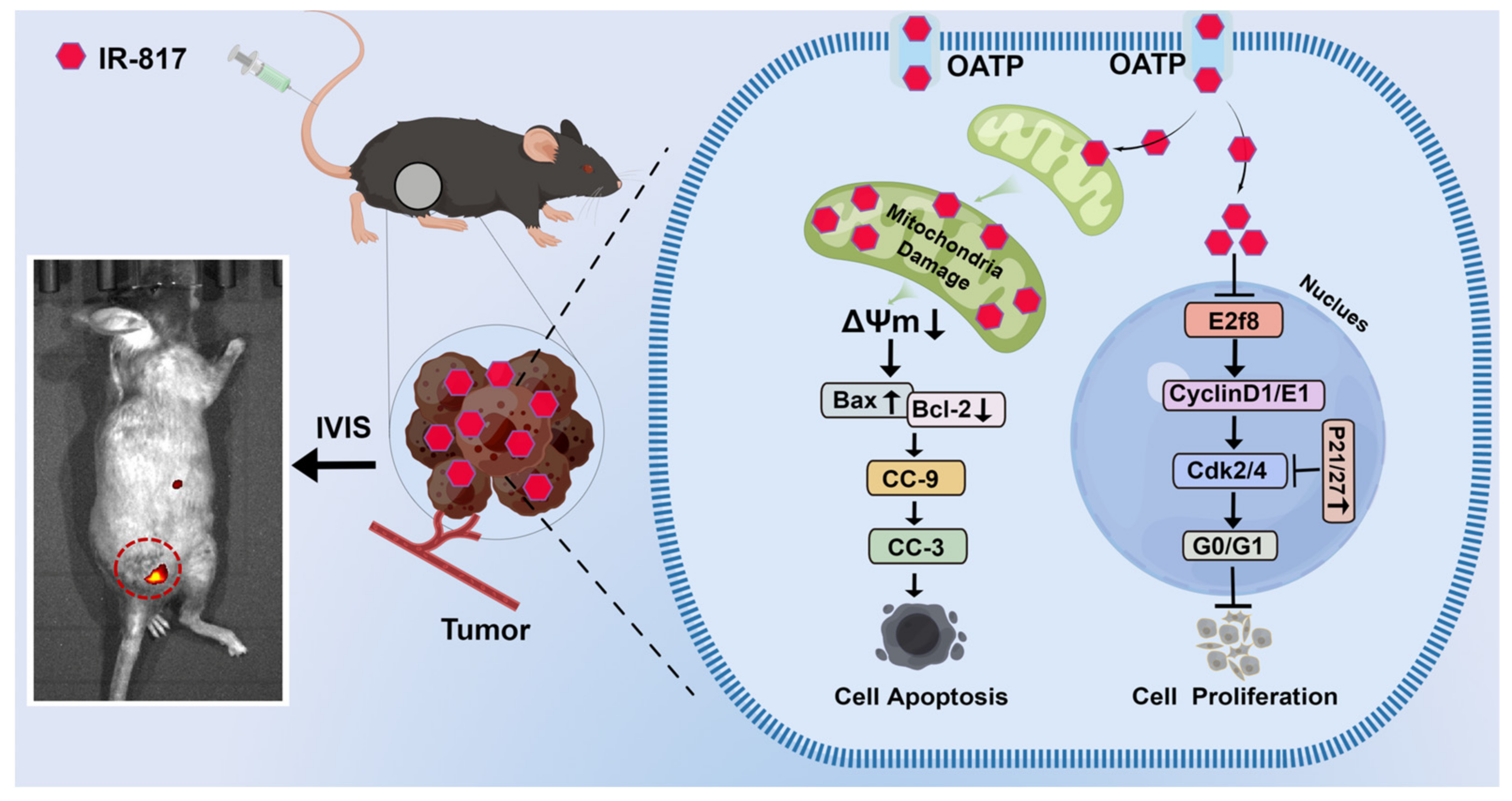
2.3. IR-817 Specifically Targeted Mitochondria and Entered Cells through OATP Transporters
2.4. IR-817 Induced Apoptosis of Melanoma Cells A375 and B16-F10 through the Mitochondrial Apoptotic Pathway
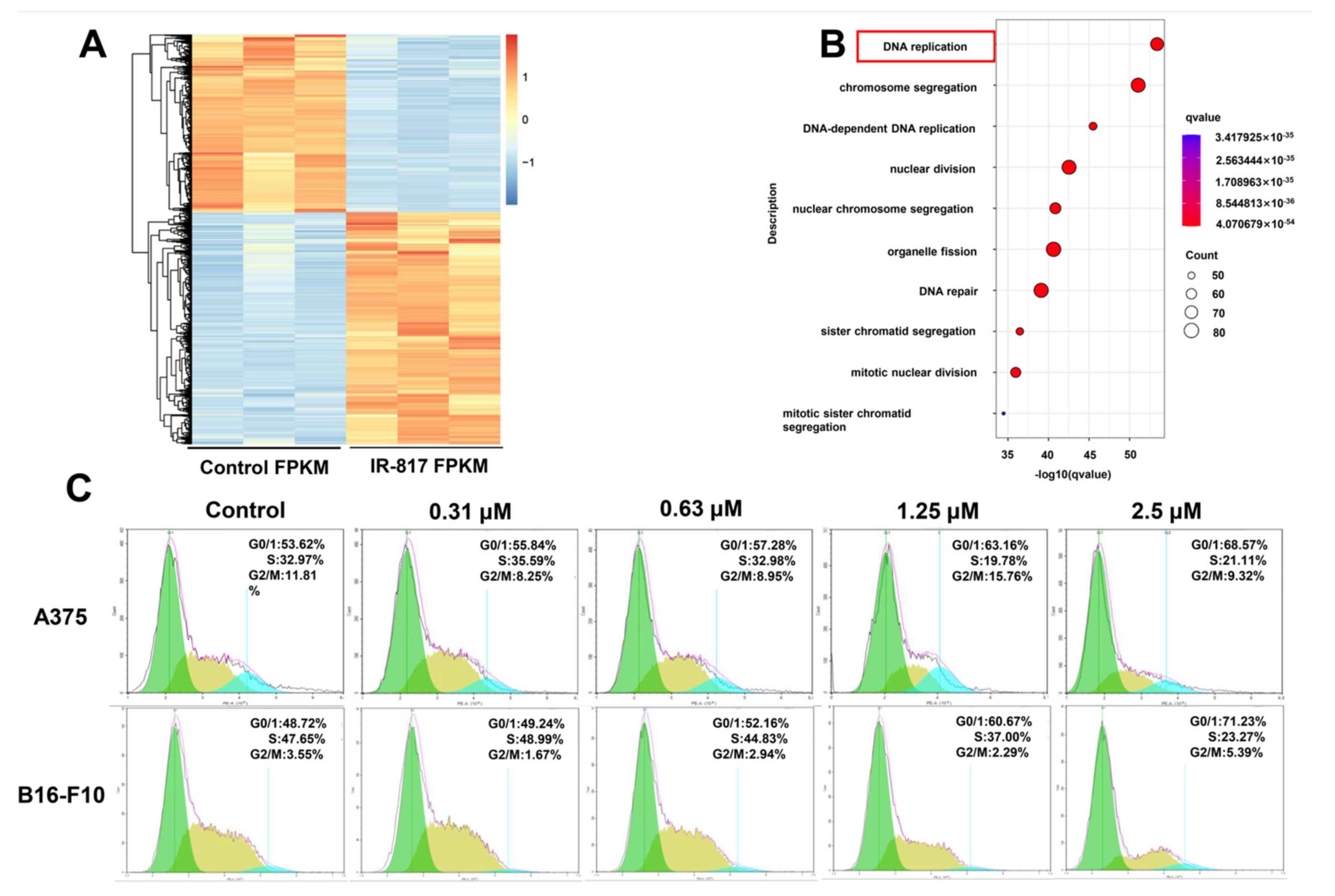
2.5. IR-817 Selectively Inhibited Melanoma Cell Proliferation
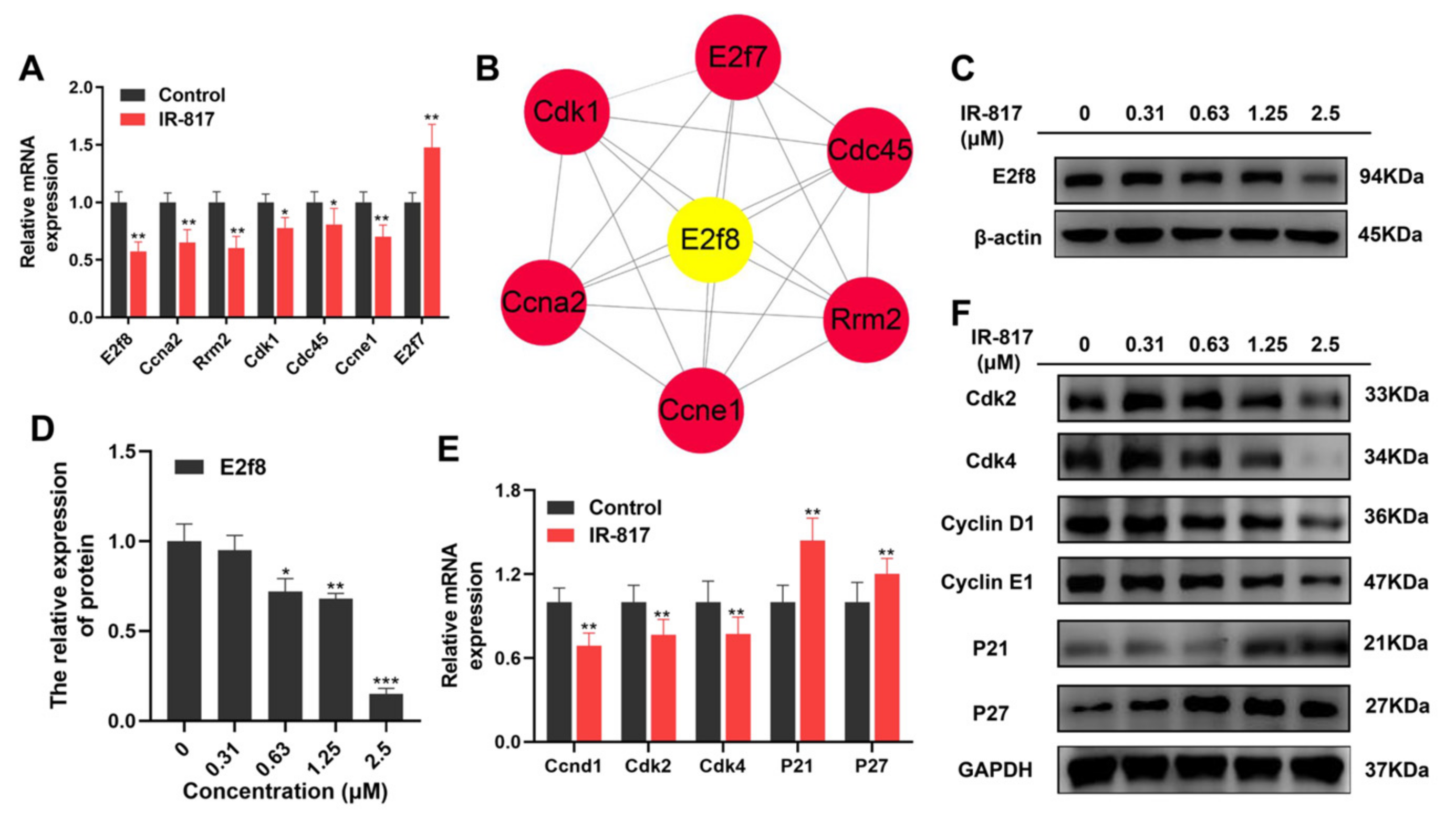
2.6. IR-817 Induced Melanoma Cell Cycle Arrest at the G0/G1 Phase by E2F/Cyclin/CDK Signal Regulation Network
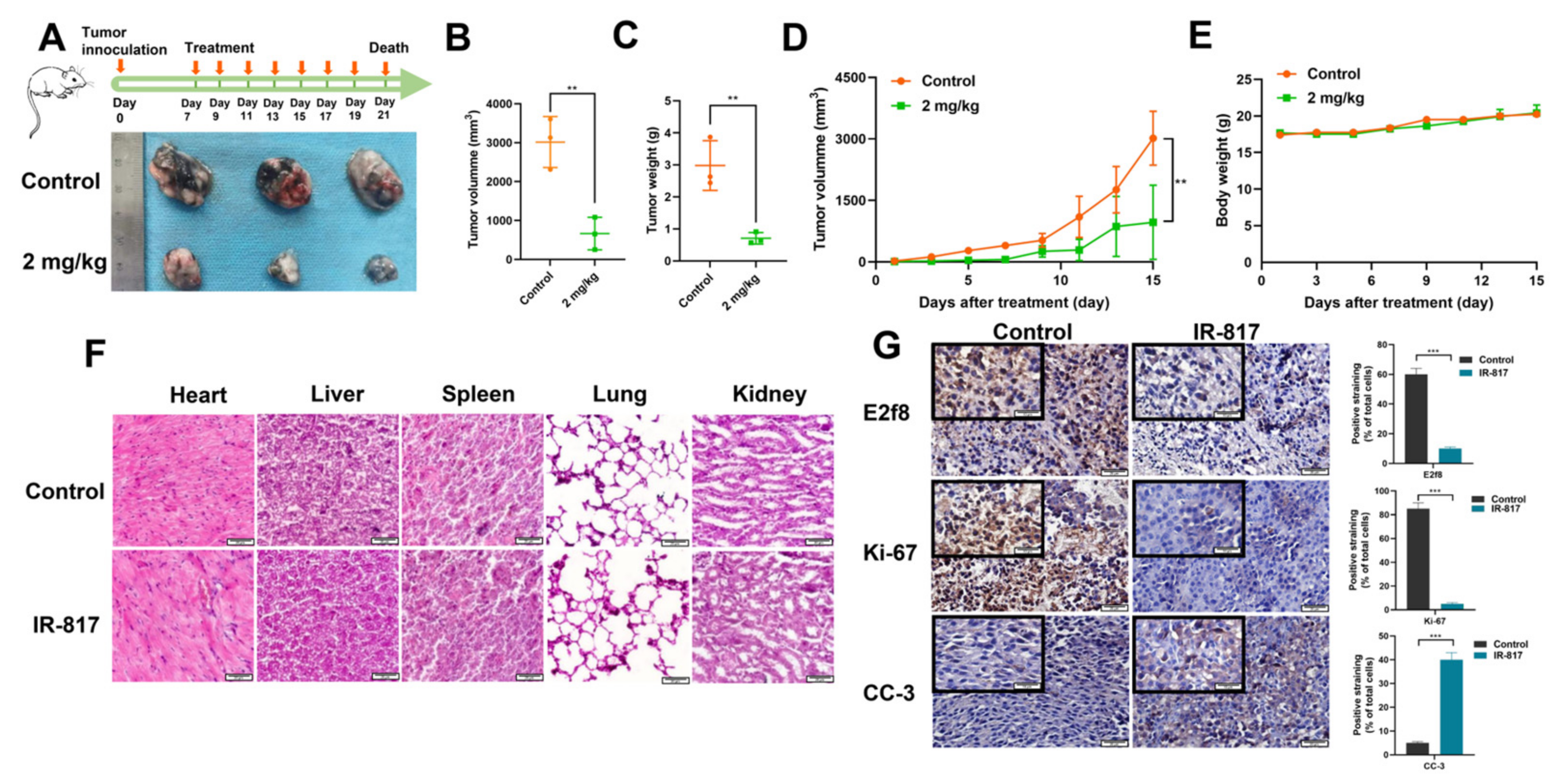
2.7. IR-817 Inhibited the Growth of Melanoma In Vivo
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Synthesis of IR-817
4.3. Cell Culture
4.4. Animal Models
4.5. Fluorescence Optical Properties
4.6. Intracellular Imaging
4.7. Fluorescence Imaging In Vivo
4.8. Imaging of Subcellular Localization
4.9. The Cellular Uptake Mechanism of IR-817
4.10. Cells Apoptosis Assay
4.11. Mitochondrial Membrane Potential (ΔΨm) Detection Assay
4.12. Western Blotting
4.13. Anti-Proliferation Assay In Vitro
4.14. RNA Sequence and Analysis
4.15. Quantitative Real-Time Reverse Transcript PCR (qRT-PCR)
4.16. Cell Cycle Assay
4.17. Anti-Tumor Assays In Vivo
4.18. Biodistribution of IR-817 in Tissues
4.19. Hematoxylin and Eosin (H&E) Staining
4.20. Immunohistochemistry
4.21. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laikova, K.V.; Oberemok, V.V.; Krasnodubets, A.M.; Gal’chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Temirova, Z.Z.; Gorlov, M.V.; Shved, N.A.; Kumeiko, V.V.; et al. Advances in the Understanding of Skin Cancer: Ultraviolet Radiationtations, and Antisense Oligonucleotides as Anticancer Drugs. Molecules 2019, 24, 1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.H.; Lee, K.J.; Chiu, Y.H.; Huang, K.C.; Wang, G.S.; Chen, L.P.; Liao, K.W.; Lin, C.S. The Lysosome in Malignant Melanoma: Biology, Function and Therapeutic Applications. Cells 2022, 11, 1492. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, S.S.; Qin, X.; Zhou, J.; Li, L.; Huang, W.; Yao, S.Q. Smart Design of Nanomaterials for Mitochondria-Targeted Nanotherapeutics. Angew. Chem. Int. Ed. Engl. 2021, 60, 2232–2256. [Google Scholar] [CrossRef] [PubMed]
- Buchke, S.; Sharma, M.; Bora, A.; Relekar, M.; Bhanu, P.; Kumar, J. Mitochondria-Targeted, Nanoparticle-Based Drug-Delivery Systems: Therapeutics for Mitochondrial Disorders. Life 2022, 12, 657. [Google Scholar] [CrossRef]
- Huang, M.; Myers, C.R.; Wang, Y.; You, M. Mitochondria as a Novel Target for Cancer Chemoprevention: Emergence of Mitochondrial-targeting Agents. Cancer Prev. Res. 2021, 14, 285–306. [Google Scholar] [CrossRef]
- Gao, M.; Huang, X.; Wu, Z.; Wang, L.; Yuan, S.; Du, Z.; Luo, S.; Li, R.; Wang, W. Synthesis of a versatile mitochondria-targeting small molecule for cancer near-infrared fluorescent imaging and radio/photodynamic/photothermal synergistic therapies. Mater. Today Bio. 2022, 15, 100316. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, W.; Kuang, X.; Hou, S.; Liu, H. Nanopreparations for mitochondria targeting drug delivery system: Current strategies and future prospective. Asian J. Pharm. Sci. 2017, 12, 498–508. [Google Scholar] [CrossRef]
- Guo, X.; Yang, N.; Ji, W.; Zhang, H.; Dong, X.; Zhou, Z.; Li, L.; Shen, H.M.; Yao, S.Q.; Huang, W. Mito-Bomb: Targeting Mitochondria for Cancer Therapy. Adv. Mater. 2021, 33, e2007778. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.; Ma, H.; Wang, H.; Zhang, R.; Zhang, X.D. Activatable NIR-II organic fluorescent probes for bioimaging. Theranostics 2022, 12, 3345–3371. [Google Scholar] [CrossRef] [PubMed]
- Usama, S.M.; Zhao, B.; Burgess, K. A Near-IR Fluorescent Dasatinib Derivative That Localizes in Cancer Cells. Bioconjug. Chem. 2019, 30, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.B.; Shi, C.; Chu, G.C.; Xu, Q.; Zhang, Y.; Li, Q.; Yu, J.S.; Zhau, H.E.; Chung, L.W. Near-infrared fluorescence heptamethine carbocyanine dyes mediate imaging and targeted drug delivery for human brain tumor. Biomaterials 2015, 67, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Luo, S.; Long, L.; Wang, Y.; Wang, D.; Fang, S.; Ouyang, Q.; Su, Y.; Cheng, T.; Shi, C. Structure-Guided Design and Synthesis of a Mitochondria-Targeting Near-Infrared Fluorophore with Multimodal Therapeutic Activities. Adv. Mater. 2017, 29, 1704196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, S.; Zhang, C.; Liao, X.; Liu, T.; Jiang, Z.; Liu, D.; Tan, X.; Long, L.; Wang, Y.; et al. An NIR-Fluorophore-Based Therapeutic Endoplasmic Reticulum Stress Inducer. Adv. Mater. 2018, 30, 1800475. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zeng, Z.; Shan, T.; Xu, X.; Chen, J.; He, Y.; Zhang, T.; Huang, Z.; Chai, G.; Huang, Y.; et al. Fibroblast activation protein α activatable theranostic pro-photosensitizer for accurate tumor imaging and highly-specific photodynamic therapy. Theranostics 2022, 12, 3610–3627. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Y.; Yue, X.; Dai, Z. Cyanine conjugates in cancer theranostics. Bioact. Mater. 2021, 6, 794–809. [Google Scholar] [CrossRef]
- Lim, W.; Byun, J.Y.; Jo, G.; Kim, E.J.; Park, M.H.; Hyun, H. Molecular Tuning of IR-786 for Improved Tumor Imaging and Photothermal Therapy. Pharmaceutics 2022, 14, 676. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Sun, C.; He, Y.; Xia, T.; Wang, J.; Xiong, X.; Zhang, Q.; Yang, S.; Liu, L. ZWZ-3, a Fluorescent Probe Targeting Mitochondria for Melanoma Imaging and Therapy. Front. Pharmacol. 2022, 13, 829684. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, T.; Su, Y.; Luo, S.; Zhu, Y.; Tan, X.; Fan, S.; Zhang, L.; Zhou, Y.; Cheng, T.; et al. A near-infrared fluorescent heptamethine indocyanine dye with preferential tumor accumulation for in vivo imaging. Biomaterials 2010, 31, 6612–6617. [Google Scholar]
- Zhang, T.; Wu, B.; Akakuru, O.U.; Yao, C.; Sun, S.; Chen, L.; Ren, W.; Wu, A.; Huang, P. Hsp90 inhibitor-loaded IR780 micelles for mitochondria-targeted mild-temperature photothermal therapy in xenograft models of human breast cancer. Cancer Lett. 2021, 500, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Du, W.; Zhao, Y.; Lim, K.; Lu, L.; Zhang, C.; Li, L. Mitochondria targeting drugs for neurodegenerative diseases-Design, mechanism and application. Acta Pharm. Sin. B 2022, 12, 2778–2789. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhou, J.; Luo, S.; Wang, Y.; He, J.; Luo, P.; Chen, Z.; Liu, T.; Tan, X.; Ou, J.; et al. Identification of a fluorescent small-molecule enhancer for therapeutic autophagy in colorectal cancer by targeting mitochondrial protein translocase TIM44. Gut 2018, 67, 307–319. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, J.; Lu, W. The Significance of Mitochondrial Dysfunction in Cancer. Int. J. Mol. Sci. 2020, 21, 5598. [Google Scholar] [CrossRef] [PubMed]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Tian, M.; Ma, Y.; Lin, W. Fluorescent Probes for the Visualization of Cell Viability. Acc. Chem. Res. 2019, 52, 2147–2157. [Google Scholar] [CrossRef]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe. Bio. Protoc. 2019, 9, e3128. [Google Scholar] [CrossRef]
- Wu, T.; Geng, J.; Guo, W.; Gao, J.; Zhu, X. Asiatic acid inhibits lung cancer cell growth in vitro and in vivo by destroying mitochondria. Acta Pharm. Sin. B 2017, 7, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Wang, X.; Weng, Y.H.; Jin, X.; Ji, J.L.; Guo, L.; Hu, B.; Liu, N.; Cheng, Q.; Zhang, J.; et al. siRNA Knockdown of RRM2 Effectively Suppressed Pancreatic Tumor Growth Alone or Synergistically with Doxorubicin. Mol. Ther. Nucleic Acids 2018, 12, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, J.; Huang, Y.; Peng, W.; Qian, W.; Gu, J.; Wang, Q.; Hu, T.; Ji, D.; Ji, B.; et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif. 2018, 51, e12505. [Google Scholar] [CrossRef] [Green Version]
- Parisi, F.; Ariyan, S.; Narayan, D.; Bacchiocchi, A.; Hoyt, K.; Cheng, E.; Xu, F.; Li, P.; Halaban, R.; Kluger, Y. Detecting copy number status and uncovering subclonal markers in heterogeneous tumor biopsies. BMC Genom. 2011, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Targen, S.; Konu, O. Zebrafish Xenotransplantation Models for Studying Gene Function and Drug Treatment in Hepatocellular Carcinoma. J. Gastrointest. Cancer 2021, 52, 1248–1265. [Google Scholar] [CrossRef] [PubMed]
- van Rooijen, E.; Fazio, M.; Zon, L.I. From fish bowl to bedside: The power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics. Pigment Cell Melanoma Res. 2017, 30, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J. How to unleash mitochondrial apoptotic blockades to kill cancers? Acta. Pharm. Sin. B 2017, 7, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Birkinshaw, R.W.; Czabotar, P.E. The BCL-2 family of proteins and mitochondrial outer membrane permeabilisation. Semin. Cell Dev. Biol. 2017, 72, 152–162. [Google Scholar] [CrossRef]
- Peña-Blanco, A.; García-Sáez, A.J. Bax, Bak and beyond—Mitochondrial performance in apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R. The Mitochondrial Pathway of Apoptosis Part II: The BCL-2 Protein Family. Cold Spring Harb Perspect. Biol. 2022, 14, a041046. [Google Scholar] [CrossRef]
- Kim, L.K.; Park, S.A.; Eoh, K.J.; Heo, T.H.; Kim, Y.T.; Kim, H.J. E2F8 regulates the proliferation and invasion through epithelial-mesenchymal transition in cervical cancer. Int. J. Biol. Sci. 2020, 16, 320–329. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Christensen, J.; Cloos, P.; Toftegaard, U.; Klinkenberg, D.; Bracken, A.P.; Trinh, E.; Heeran, M.; Di Stefano, L.; Helin, K. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res 2005, 33, 5458–5470. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Xiao, J.; Liu, J.; Xing, F. E2F8 is a Potential Therapeutic Target for Hepatocellular Carcinoma. J. Cancer 2017, 8, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.J.; Enrico, T.P.; Mouery, R.D.; Wasserman, D.; Nachum, S.; Tzur, A. Complex Cartography: Regulation of E2F Transcription Factors by Cyclin F and Ubiquitin. Trends Cell Biol. 2020, 30, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.K.; Shah, M.A. Targeting the cell cycle: A new approach to cancer therapy. J. Clin. Oncol. 2005, 23, 9408–9421. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; You, J.; Wu, Q.; Meng, W.; He, Q.; Yang, B.; Zhu, C.; Cao, J. Cyclin-dependent kinases-based synthetic lethality: Evidence, concept, and strategy. Acta Pharm. Sin. B 2021, 11, 2738–2748. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Xu, F.; Xiong, M.; Yang, H.; Lin, W.; Xie, Y.; Xi, H.; Xue, Q.; Ye, T.; Yu, L. Repurposing of antipsychotic trifluoperazine for treating brain metastasis, lung metastasis and bone metastasis of melanoma by disrupting autophagy flux. Pharmacol. Res. 2021, 163, 105295. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Luo, S.; Wang, D.; Su, Y.; Cheng, T.; Shi, C. A NIR heptamethine dye with intrinsic cancer targeting, imaging and photosensitizing properties. Biomaterials 2012, 33, 2230–2239. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Tan, X.; Qi, Q.; Guo, Q.; Ran, X.; Zhang, L.; Zhang, E.; Liang, Y.; Weng, L.; Zheng, H.; et al. A multifunctional heptamethine near-infrared dye for cancer theranosis. Biomaterials 2013, 34, 2244–2251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, W.; Li, B.; Li, W.; Zhang, C.; Hou, X.; Jiang, J.; Dong, Y. Ratiometric fluorescent probes for capturing endogenous hypochlorous acid in the lungs of mice. Chem. Sci. 2018, 9, 8207–8212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chen, L.; Zhang, H.; Huang, S.; Xiong, Y.; Xia, C. Interaction of Sulfonylureas with Liver Uptake Transporters OATP1B1 and OATP1B3. Basic Clin. Pharmacol. Toxicol. 2018, 123, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Li, J.; Jing, X.; Ding, X.; Yu, Y.; Zhao, Q. Fucoidan Induces Apoptosis and Inhibits Proliferation of Hepatocellular Carcinoma via the p38 MAPK/ERK and PI3K/Akt Signal Pathways. Cancer Manag. Res. 2020, 12, 1713–1723. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Wang, J.; Xia, T.; Sun, Q.; He, Y.; Wang, H.; He, Q.; Liu, L. Mitochondrion-Targeted NIR Therapeutic Agent Suppresses Melanoma by Inducing Apoptosis and Cell Cycle Arrest via E2F/Cyclin/CDK Pathway. Pharmaceuticals 2022, 15, 1589. https://doi.org/10.3390/ph15121589
Sun C, Wang J, Xia T, Sun Q, He Y, Wang H, He Q, Liu L. Mitochondrion-Targeted NIR Therapeutic Agent Suppresses Melanoma by Inducing Apoptosis and Cell Cycle Arrest via E2F/Cyclin/CDK Pathway. Pharmaceuticals. 2022; 15(12):1589. https://doi.org/10.3390/ph15121589
Chicago/Turabian StyleSun, Changzhen, Jianv Wang, Tong Xia, Qin Sun, Yijing He, Hailan Wang, Qizhou He, and Li Liu. 2022. "Mitochondrion-Targeted NIR Therapeutic Agent Suppresses Melanoma by Inducing Apoptosis and Cell Cycle Arrest via E2F/Cyclin/CDK Pathway" Pharmaceuticals 15, no. 12: 1589. https://doi.org/10.3390/ph15121589
APA StyleSun, C., Wang, J., Xia, T., Sun, Q., He, Y., Wang, H., He, Q., & Liu, L. (2022). Mitochondrion-Targeted NIR Therapeutic Agent Suppresses Melanoma by Inducing Apoptosis and Cell Cycle Arrest via E2F/Cyclin/CDK Pathway. Pharmaceuticals, 15(12), 1589. https://doi.org/10.3390/ph15121589




